Junru Cheng
Towards a Comprehensive Benchmark for Pathological Lymph Node Metastasis in Breast Cancer Sections
Nov 16, 2024Abstract:Advances in optical microscopy scanning have significantly contributed to computational pathology (CPath) by converting traditional histopathological slides into whole slide images (WSIs). This development enables comprehensive digital reviews by pathologists and accelerates AI-driven diagnostic support for WSI analysis. Recent advances in foundational pathology models have increased the need for benchmarking tasks. The Camelyon series is one of the most widely used open-source datasets in computational pathology. However, the quality, accessibility, and clinical relevance of the labels have not been comprehensively evaluated. In this study, we reprocessed 1,399 WSIs and labels from the Camelyon-16 and Camelyon-17 datasets, removing low-quality slides, correcting erroneous labels, and providing expert pixel annotations for tumor regions in the previously unreleased test set. Based on the sizes of re-annotated tumor regions, we upgraded the binary cancer screening task to a four-class task: negative, micro-metastasis, macro-metastasis, and Isolated Tumor Cells (ITC). We reevaluated pre-trained pathology feature extractors and multiple instance learning (MIL) methods using the cleaned dataset, providing a benchmark that advances AI development in histopathology.
Agent Aggregator with Mask Denoise Mechanism for Histopathology Whole Slide Image Analysis
Sep 18, 2024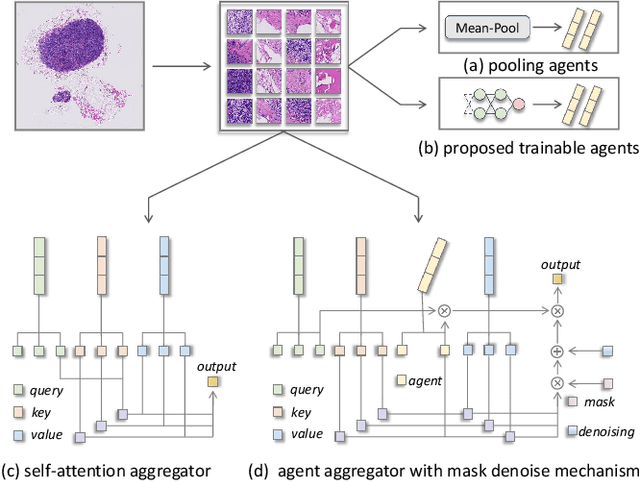
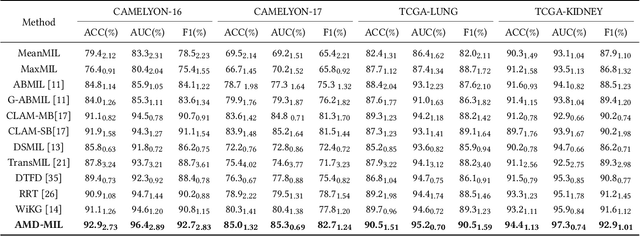
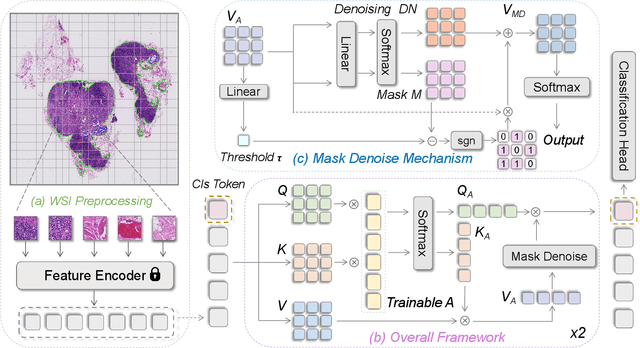
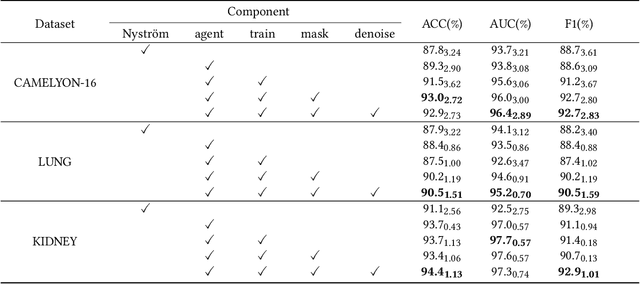
Abstract:Histopathology analysis is the gold standard for medical diagnosis. Accurate classification of whole slide images (WSIs) and region-of-interests (ROIs) localization can assist pathologists in diagnosis. The gigapixel resolution of WSI and the absence of fine-grained annotations make direct classification and analysis challenging. In weakly supervised learning, multiple instance learning (MIL) presents a promising approach for WSI classification. The prevailing strategy is to use attention mechanisms to measure instance importance for classification. However, attention mechanisms fail to capture inter-instance information, and self-attention causes quadratic computational complexity. To address these challenges, we propose AMD-MIL, an agent aggregator with a mask denoise mechanism. The agent token acts as an intermediate variable between the query and key for computing instance importance. Mask and denoising matrices, mapped from agents-aggregated value, dynamically mask low-contribution representations and eliminate noise. AMD-MIL achieves better attention allocation by adjusting feature representations, capturing micro-metastases in cancer, and improving interpretability. Extensive experiments on CAMELYON-16, CAMELYON-17, TCGA-KIDNEY, and TCGA-LUNG show AMD-MIL's superiority over state-of-the-art methods.
The Whole Pathological Slide Classification via Weakly Supervised Learning
Jul 12, 2023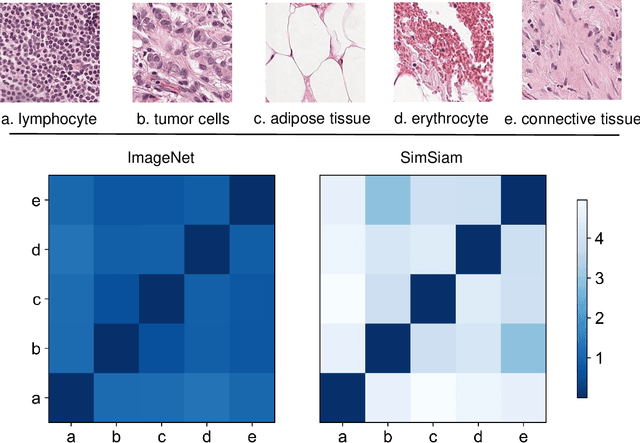
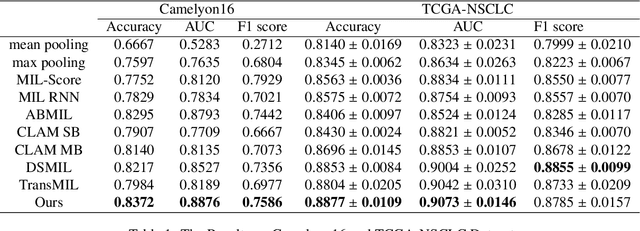
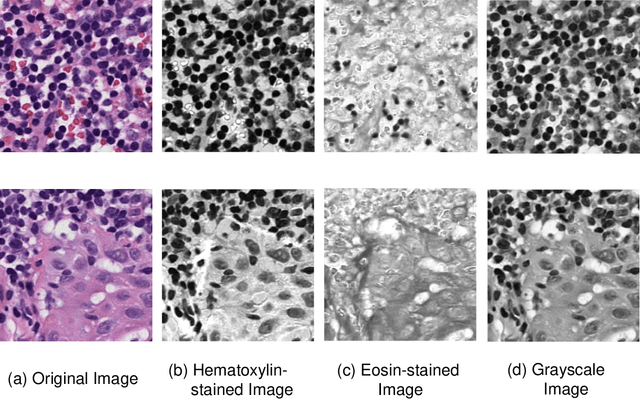
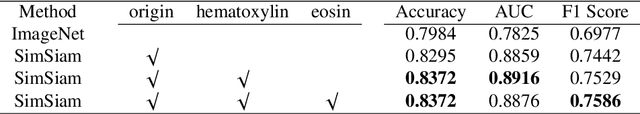
Abstract:Due to its superior efficiency in utilizing annotations and addressing gigapixel-sized images, multiple instance learning (MIL) has shown great promise as a framework for whole slide image (WSI) classification in digital pathology diagnosis. However, existing methods tend to focus on advanced aggregators with different structures, often overlooking the intrinsic features of H\&E pathological slides. To address this limitation, we introduced two pathological priors: nuclear heterogeneity of diseased cells and spatial correlation of pathological tiles. Leveraging the former, we proposed a data augmentation method that utilizes stain separation during extractor training via a contrastive learning strategy to obtain instance-level representations. We then described the spatial relationships between the tiles using an adjacency matrix. By integrating these two views, we designed a multi-instance framework for analyzing H\&E-stained tissue images based on pathological inductive bias, encompassing feature extraction, filtering, and aggregation. Extensive experiments on the Camelyon16 breast dataset and TCGA-NSCLC Lung dataset demonstrate that our proposed framework can effectively handle tasks related to cancer detection and differentiation of subtypes, outperforming state-of-the-art medical image classification methods based on MIL. The code will be released later.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge