Felix Gonda
VICE: Visual Identification and Correction of Neural Circuit Errors
May 14, 2021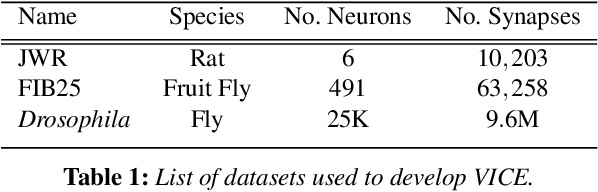
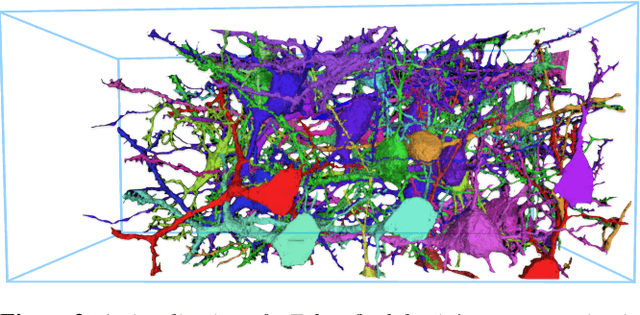
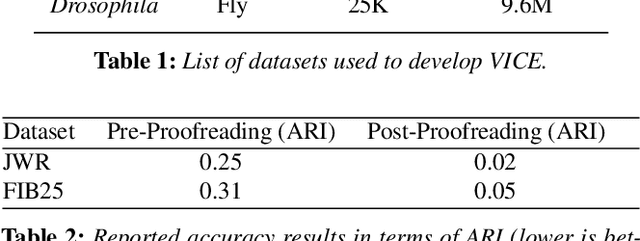
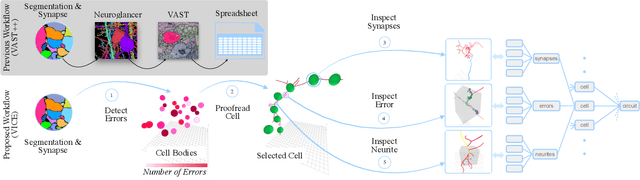
Abstract:A connectivity graph of neurons at the resolution of single synapses provides scientists with a tool for understanding the nervous system in health and disease. Recent advances in automatic image segmentation and synapse prediction in electron microscopy (EM) datasets of the brain have made reconstructions of neurons possible at the nanometer scale. However, automatic segmentation sometimes struggles to segment large neurons correctly, requiring human effort to proofread its output. General proofreading involves inspecting large volumes to correct segmentation errors at the pixel level, a visually intensive and time-consuming process. This paper presents the design and implementation of an analytics framework that streamlines proofreading, focusing on connectivity-related errors. We accomplish this with automated likely-error detection and synapse clustering that drives the proofreading effort with highly interactive 3D visualizations. In particular, our strategy centers on proofreading the local circuit of a single cell to ensure a basic level of completeness. We demonstrate our framework's utility with a user study and report quantitative and subjective feedback from our users. Overall, users find the framework more efficient for proofreading, understanding evolving graphs, and sharing error correction strategies.
Consistent Recurrent Neural Networks for 3D Neuron Segmentation
Feb 01, 2021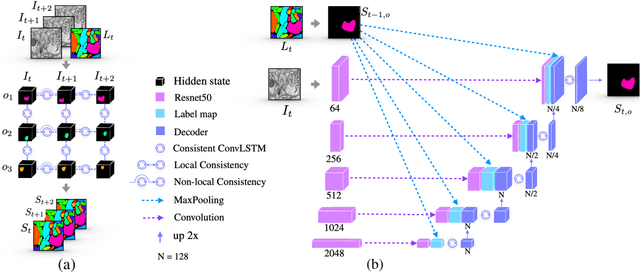

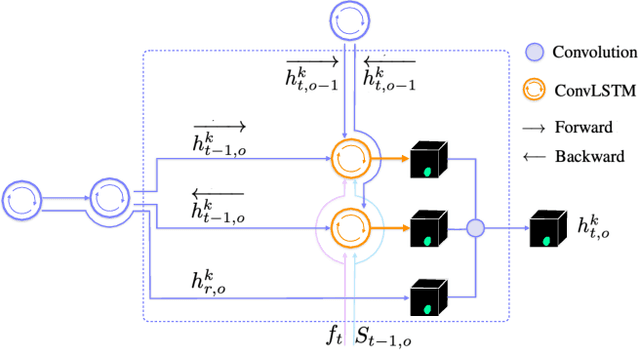

Abstract:We present a recurrent network for the 3D reconstruction of neurons that sequentially generates binary masks for every object in an image with spatio-temporal consistency. Our network models consistency in two parts: (i) local, which allows exploring non-occluding and temporally-adjacent object relationships with bi-directional recurrence. (ii) non-local, which allows exploring long-range object relationships in the temporal domain with skip connections. Our proposed network is end-to-end trainable from an input image to a sequence of object masks, and, compared to methods relying on object boundaries, its output does not require post-processing. We evaluate our method on three benchmarks for neuron segmentation and achieved state-of-the-art performance on the SNEMI3D challenge.
Parallel Separable 3D Convolution for Video and Volumetric Data Understanding
Sep 11, 2018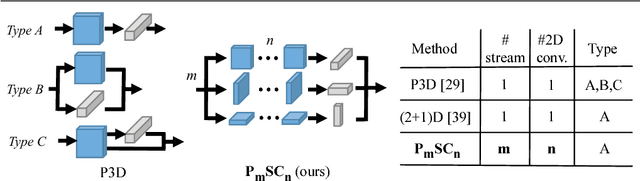
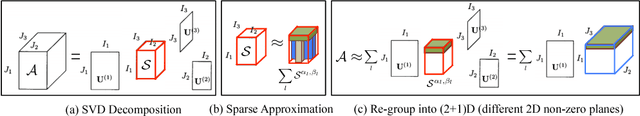
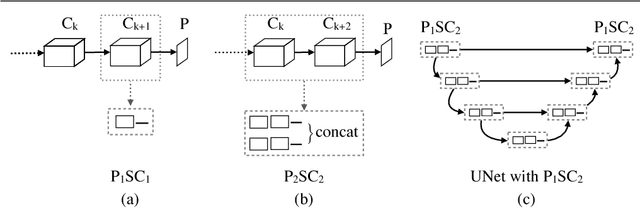
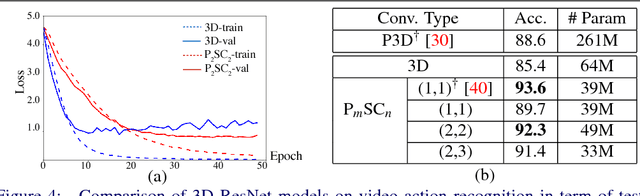
Abstract:For video and volumetric data understanding, 3D convolution layers are widely used in deep learning, however, at the cost of increasing computation and training time. Recent works seek to replace the 3D convolution layer with convolution blocks, e.g. structured combinations of 2D and 1D convolution layers. In this paper, we propose a novel convolution block, Parallel Separable 3D Convolution (PmSCn), which applies m parallel streams of n 2D and one 1D convolution layers along different dimensions. We first mathematically justify the need of parallel streams (Pm) to replace a single 3D convolution layer through tensor decomposition. Then we jointly replace consecutive 3D convolution layers, common in modern network architectures, with the multiple 2D convolution layers (Cn). Lastly, we empirically show that PmSCn is applicable to different backbone architectures, such as ResNet, DenseNet, and UNet, for different applications, such as video action recognition, MRI brain segmentation, and electron microscopy segmentation. In all three applications, we replace the 3D convolution layers in state-of-the art models with PmSCn and achieve around 14% improvement in test performance and 40% reduction in model size and on average.
Icon: An Interactive Approach to Train Deep Neural Networks for Segmentation of Neuronal Structures
Oct 27, 2016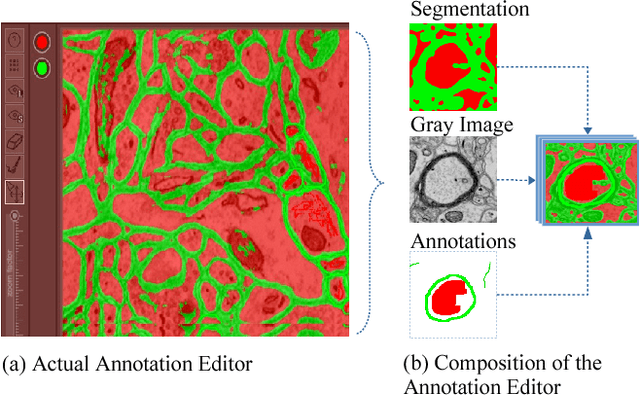
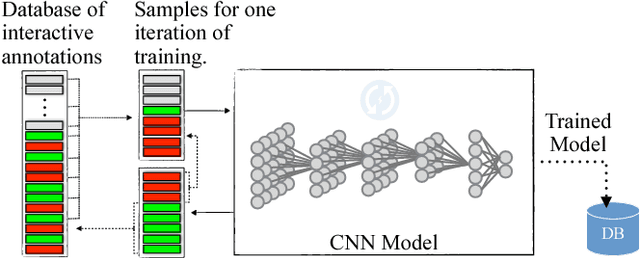
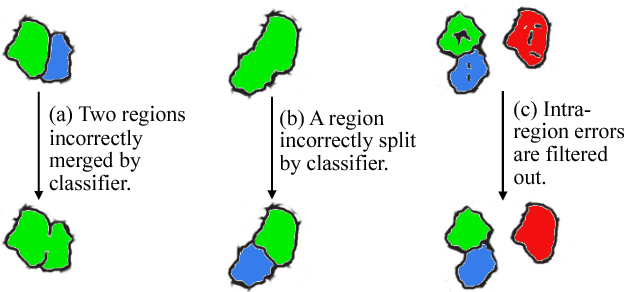
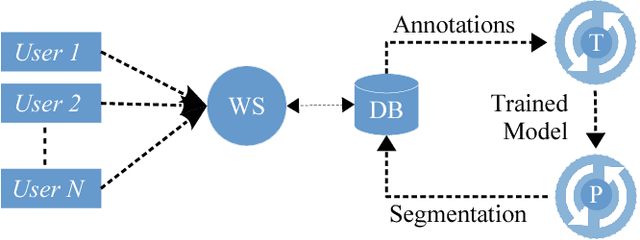
Abstract:We present an interactive approach to train a deep neural network pixel classifier for the segmentation of neuronal structures. An interactive training scheme reduces the extremely tedious manual annotation task that is typically required for deep networks to perform well on image segmentation problems. Our proposed method employs a feedback loop that captures sparse annotations using a graphical user interface, trains a deep neural network based on recent and past annotations, and displays the prediction output to users in almost real-time. Our implementation of the algorithm also allows multiple users to provide annotations in parallel and receive feedback from the same classifier. Quick feedback on classifier performance in an interactive setting enables users to identify and label examples that are more important than others for segmentation purposes. Our experiments show that an interactively-trained pixel classifier produces better region segmentation results on Electron Microscopy (EM) images than those generated by a network of the same architecture trained offline on exhaustive ground-truth labels.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge