David Clunie
In search of truth: Evaluating concordance of AI-based anatomy segmentation models
Dec 17, 2025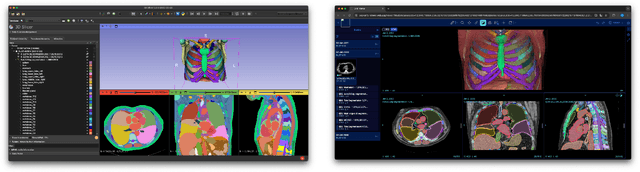


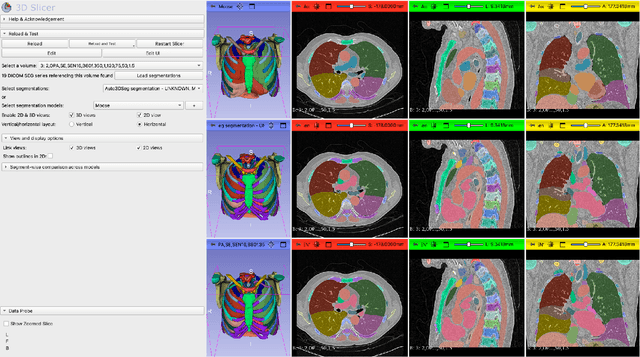
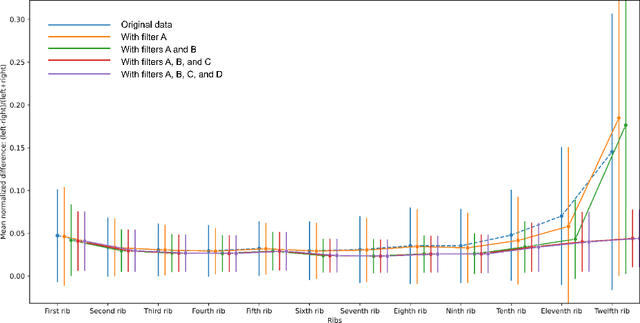
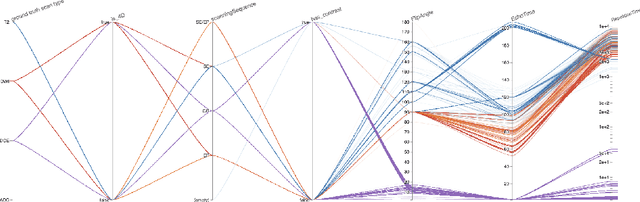
Abstract:Purpose AI-based methods for anatomy segmentation can help automate characterization of large imaging datasets. The growing number of similar in functionality models raises the challenge of evaluating them on datasets that do not contain ground truth annotations. We introduce a practical framework to assist in this task. Approach We harmonize the segmentation results into a standard, interoperable representation, which enables consistent, terminology-based labeling of the structures. We extend 3D Slicer to streamline loading and comparison of these harmonized segmentations, and demonstrate how standard representation simplifies review of the results using interactive summary plots and browser-based visualization using OHIF Viewer. To demonstrate the utility of the approach we apply it to evaluating segmentation of 31 anatomical structures (lungs, vertebrae, ribs, and heart) by six open-source models - TotalSegmentator 1.5 and 2.6, Auto3DSeg, MOOSE, MultiTalent, and CADS - for a sample of Computed Tomography (CT) scans from the publicly available National Lung Screening Trial (NLST) dataset. Results We demonstrate the utility of the framework in enabling automating loading, structure-wise inspection and comparison across models. Preliminary results ascertain practical utility of the approach in allowing quick detection and review of problematic results. The comparison shows excellent agreement segmenting some (e.g., lung) but not all structures (e.g., some models produce invalid vertebrae or rib segmentations). Conclusions The resources developed are linked from https://imagingdatacommons.github.io/segmentation-comparison/ including segmentation harmonization scripts, summary plots, and visualization tools. This work assists in model evaluation in absence of ground truth, ultimately enabling informed model selection.
Unlocking the Potential of Digital Pathology: Novel Baselines for Compression
Dec 17, 2024



Abstract:Digital pathology offers a groundbreaking opportunity to transform clinical practice in histopathological image analysis, yet faces a significant hurdle: the substantial file sizes of pathological Whole Slide Images (WSI). While current digital pathology solutions rely on lossy JPEG compression to address this issue, lossy compression can introduce color and texture disparities, potentially impacting clinical decision-making. While prior research addresses perceptual image quality and downstream performance independently of each other, we jointly evaluate compression schemes for perceptual and downstream task quality on four different datasets. In addition, we collect an initially uncompressed dataset for an unbiased perceptual evaluation of compression schemes. Our results show that deep learning models fine-tuned for perceptual quality outperform conventional compression schemes like JPEG-XL or WebP for further compression of WSI. However, they exhibit a significant bias towards the compression artifacts present in the training data and struggle to generalize across various compression schemes. We introduce a novel evaluation metric based on feature similarity between original files and compressed files that aligns very well with the actual downstream performance on the compressed WSI. Our metric allows for a general and standardized evaluation of lossy compression schemes and mitigates the requirement to independently assess different downstream tasks. Our study provides novel insights for the assessment of lossy compression schemes for WSI and encourages a unified evaluation of lossy compression schemes to accelerate the clinical uptake of digital pathology.
Rule-based outlier detection of AI-generated anatomy segmentations
Jun 20, 2024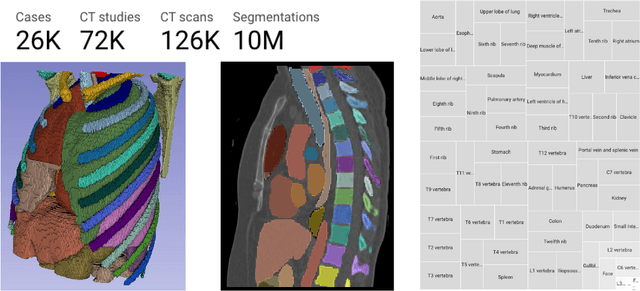
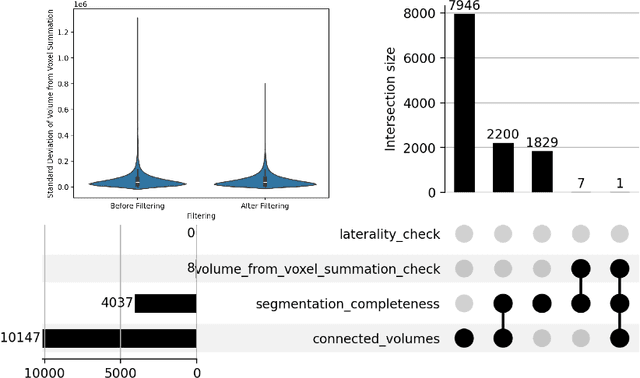
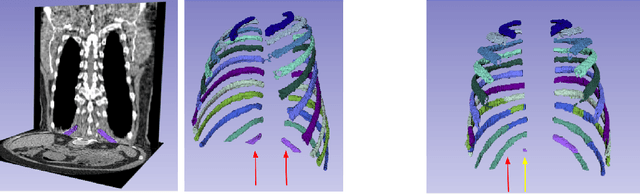
Abstract:There is a dire need for medical imaging datasets with accompanying annotations to perform downstream patient analysis. However, it is difficult to manually generate these annotations, due to the time-consuming nature, and the variability in clinical conventions. Artificial intelligence has been adopted in the field as a potential method to annotate these large datasets, however, a lack of expert annotations or ground truth can inhibit the adoption of these annotations. We recently made a dataset publicly available including annotations and extracted features of up to 104 organs for the National Lung Screening Trial using the TotalSegmentator method. However, the released dataset does not include expert-derived annotations or an assessment of the accuracy of the segmentations, limiting its usefulness. We propose the development of heuristics to assess the quality of the segmentations, providing methods to measure the consistency of the annotations and a comparison of results to the literature. We make our code and related materials publicly available at https://github.com/ImagingDataCommons/CloudSegmentatorResults and interactive tools at https://huggingface.co/spaces/ImagingDataCommons/CloudSegmentatorResults.
Automatic classification of prostate MR series type using image content and metadata
Apr 16, 2024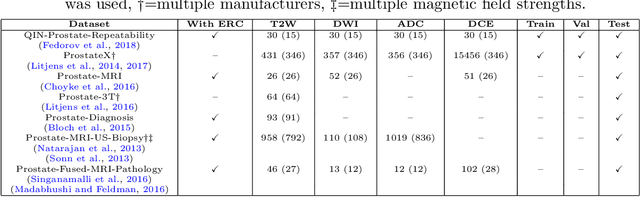
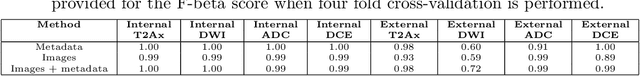
Abstract:With the wealth of medical image data, efficient curation is essential. Assigning the sequence type to magnetic resonance images is necessary for scientific studies and artificial intelligence-based analysis. However, incomplete or missing metadata prevents effective automation. We therefore propose a deep-learning method for classification of prostate cancer scanning sequences based on a combination of image data and DICOM metadata. We demonstrate superior results compared to metadata or image data alone, and make our code publicly available at https://github.com/deepakri201/DICOMScanClassification.
Enrichment of the NLST and NSCLC-Radiomics computed tomography collections with AI-derived annotations
May 31, 2023Abstract:Public imaging datasets are critical for the development and evaluation of automated tools in cancer imaging. Unfortunately, many do not include annotations or image-derived features, complicating their downstream analysis. Artificial intelligence-based annotation tools have been shown to achieve acceptable performance and thus can be used to automatically annotate large datasets. As part of the effort to enrich public data available within NCI Imaging Data Commons (IDC), here we introduce AI-generated annotations for two collections of computed tomography images of the chest, NSCLC-Radiomics, and the National Lung Screening Trial. Using publicly available AI algorithms we derived volumetric annotations of thoracic organs at risk, their corresponding radiomics features, and slice-level annotations of anatomical landmarks and regions. The resulting annotations are publicly available within IDC, where the DICOM format is used to harmonize the data and achieve FAIR principles. The annotations are accompanied by cloud-enabled notebooks demonstrating their use. This study reinforces the need for large, publicly accessible curated datasets and demonstrates how AI can be used to aid in cancer imaging.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge