Tina Kapur
Unified Cross-Modal Image Synthesis with Hierarchical Mixture of Product-of-Experts
Oct 25, 2024Abstract:We propose a deep mixture of multimodal hierarchical variational auto-encoders called MMHVAE that synthesizes missing images from observed images in different modalities. MMHVAE's design focuses on tackling four challenges: (i) creating a complex latent representation of multimodal data to generate high-resolution images; (ii) encouraging the variational distributions to estimate the missing information needed for cross-modal image synthesis; (iii) learning to fuse multimodal information in the context of missing data; (iv) leveraging dataset-level information to handle incomplete data sets at training time. Extensive experiments are performed on the challenging problem of pre-operative brain multi-parametric magnetic resonance and intra-operative ultrasound imaging.
Calibrating Expressions of Certainty
Oct 06, 2024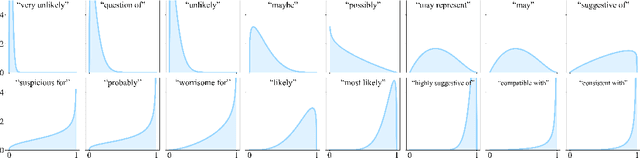
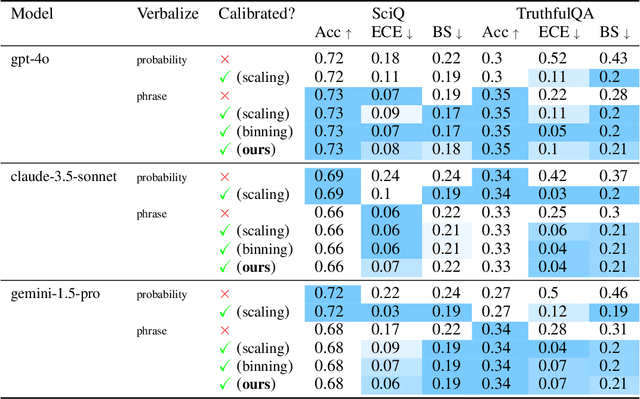
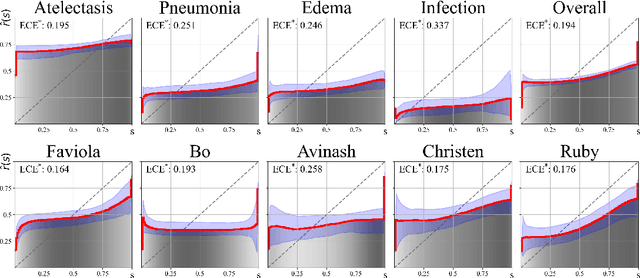
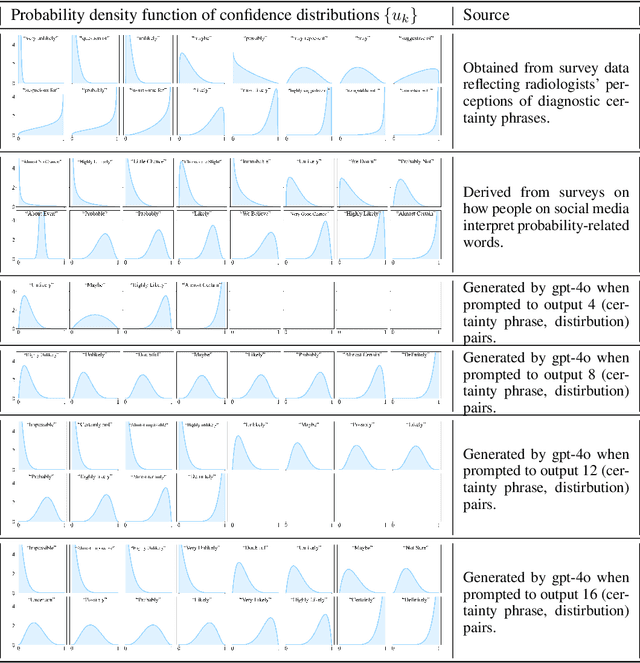
Abstract:We present a novel approach to calibrating linguistic expressions of certainty, e.g., "Maybe" and "Likely". Unlike prior work that assigns a single score to each certainty phrase, we model uncertainty as distributions over the simplex to capture their semantics more accurately. To accommodate this new representation of certainty, we generalize existing measures of miscalibration and introduce a novel post-hoc calibration method. Leveraging these tools, we analyze the calibration of both humans (e.g., radiologists) and computational models (e.g., language models) and provide interpretable suggestions to improve their calibration.
Learning to Match 2D Keypoints Across Preoperative MR and Intraoperative Ultrasound
Sep 12, 2024Abstract:We propose in this paper a texture-invariant 2D keypoints descriptor specifically designed for matching preoperative Magnetic Resonance (MR) images with intraoperative Ultrasound (US) images. We introduce a matching-by-synthesis strategy, where intraoperative US images are synthesized from MR images accounting for multiple MR modalities and intraoperative US variability. We build our training set by enforcing keypoints localization over all images then train a patient-specific descriptor network that learns texture-invariant discriminant features in a supervised contrastive manner, leading to robust keypoints descriptors. Our experiments on real cases with ground truth show the effectiveness of the proposed approach, outperforming the state-of-the-art methods and achieving 80.35% matching precision on average.
LNQ 2023 challenge: Benchmark of weakly-supervised techniques for mediastinal lymph node quantification
Aug 19, 2024



Abstract:Accurate assessment of lymph node size in 3D CT scans is crucial for cancer staging, therapeutic management, and monitoring treatment response. Existing state-of-the-art segmentation frameworks in medical imaging often rely on fully annotated datasets. However, for lymph node segmentation, these datasets are typically small due to the extensive time and expertise required to annotate the numerous lymph nodes in 3D CT scans. Weakly-supervised learning, which leverages incomplete or noisy annotations, has recently gained interest in the medical imaging community as a potential solution. Despite the variety of weakly-supervised techniques proposed, most have been validated only on private datasets or small publicly available datasets. To address this limitation, the Mediastinal Lymph Node Quantification (LNQ) challenge was organized in conjunction with the 26th International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI 2023). This challenge aimed to advance weakly-supervised segmentation methods by providing a new, partially annotated dataset and a robust evaluation framework. A total of 16 teams from 5 countries submitted predictions to the validation leaderboard, and 6 teams from 3 countries participated in the evaluation phase. The results highlighted both the potential and the current limitations of weakly-supervised approaches. On one hand, weakly-supervised approaches obtained relatively good performance with a median Dice score of $61.0\%$. On the other hand, top-ranked teams, with a median Dice score exceeding $70\%$, boosted their performance by leveraging smaller but fully annotated datasets to combine weak supervision and full supervision. This highlights both the promise of weakly-supervised methods and the ongoing need for high-quality, fully annotated data to achieve higher segmentation performance.
Patient-Specific Real-Time Segmentation in Trackerless Brain Ultrasound
May 16, 2024



Abstract:Intraoperative ultrasound (iUS) imaging has the potential to improve surgical outcomes in brain surgery. However, its interpretation is challenging, even for expert neurosurgeons. In this work, we designed the first patient-specific framework that performs brain tumor segmentation in trackerless iUS. To disambiguate ultrasound imaging and adapt to the neurosurgeon's surgical objective, a patient-specific real-time network is trained using synthetic ultrasound data generated by simulating virtual iUS sweep acquisitions in pre-operative MR data. Extensive experiments performed in real ultrasound data demonstrate the effectiveness of the proposed approach, allowing for adapting to the surgeon's definition of surgical targets and outperforming non-patient-specific models, neurosurgeon experts, and high-end tracking systems. Our code is available at: \url{https://github.com/ReubenDo/MHVAE-Seg}.
Automatic classification of prostate MR series type using image content and metadata
Apr 16, 2024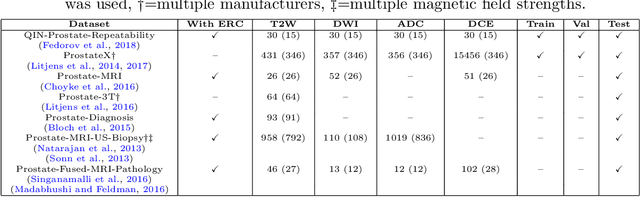
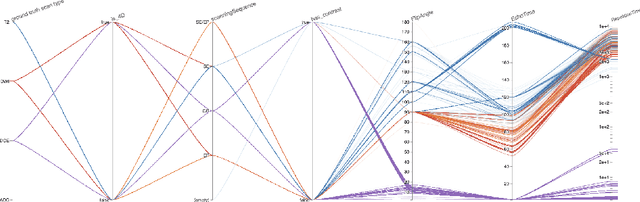
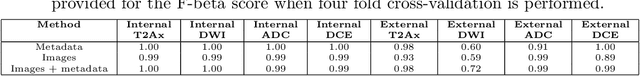
Abstract:With the wealth of medical image data, efficient curation is essential. Assigning the sequence type to magnetic resonance images is necessary for scientific studies and artificial intelligence-based analysis. However, incomplete or missing metadata prevents effective automation. We therefore propose a deep-learning method for classification of prostate cancer scanning sequences based on a combination of image data and DICOM metadata. We demonstrate superior results compared to metadata or image data alone, and make our code publicly available at https://github.com/deepakri201/DICOMScanClassification.
Learning Expected Appearances for Intraoperative Registration during Neurosurgery
Oct 03, 2023Abstract:We present a novel method for intraoperative patient-to-image registration by learning Expected Appearances. Our method uses preoperative imaging to synthesize patient-specific expected views through a surgical microscope for a predicted range of transformations. Our method estimates the camera pose by minimizing the dissimilarity between the intraoperative 2D view through the optical microscope and the synthesized expected texture. In contrast to conventional methods, our approach transfers the processing tasks to the preoperative stage, reducing thereby the impact of low-resolution, distorted, and noisy intraoperative images, that often degrade the registration accuracy. We applied our method in the context of neuronavigation during brain surgery. We evaluated our approach on synthetic data and on retrospective data from 6 clinical cases. Our method outperformed state-of-the-art methods and achieved accuracies that met current clinical standards.
Unified Brain MR-Ultrasound Synthesis using Multi-Modal Hierarchical Representations
Sep 19, 2023Abstract:We introduce MHVAE, a deep hierarchical variational auto-encoder (VAE) that synthesizes missing images from various modalities. Extending multi-modal VAEs with a hierarchical latent structure, we introduce a probabilistic formulation for fusing multi-modal images in a common latent representation while having the flexibility to handle incomplete image sets as input. Moreover, adversarial learning is employed to generate sharper images. Extensive experiments are performed on the challenging problem of joint intra-operative ultrasound (iUS) and Magnetic Resonance (MR) synthesis. Our model outperformed multi-modal VAEs, conditional GANs, and the current state-of-the-art unified method (ResViT) for synthesizing missing images, demonstrating the advantage of using a hierarchical latent representation and a principled probabilistic fusion operation. Our code is publicly available \url{https://github.com/ReubenDo/MHVAE}.
Deep Learning for Detection and Localization of B-Lines in Lung Ultrasound
Feb 15, 2023



Abstract:Lung ultrasound (LUS) is an important imaging modality used by emergency physicians to assess pulmonary congestion at the patient bedside. B-line artifacts in LUS videos are key findings associated with pulmonary congestion. Not only can the interpretation of LUS be challenging for novice operators, but visual quantification of B-lines remains subject to observer variability. In this work, we investigate the strengths and weaknesses of multiple deep learning approaches for automated B-line detection and localization in LUS videos. We curate and publish, BEDLUS, a new ultrasound dataset comprising 1,419 videos from 113 patients with a total of 15,755 expert-annotated B-lines. Based on this dataset, we present a benchmark of established deep learning methods applied to the task of B-line detection. To pave the way for interpretable quantification of B-lines, we propose a novel "single-point" approach to B-line localization using only the point of origin. Our results show that (a) the area under the receiver operating characteristic curve ranges from 0.864 to 0.955 for the benchmarked detection methods, (b) within this range, the best performance is achieved by models that leverage multiple successive frames as input, and (c) the proposed single-point approach for B-line localization reaches an F1-score of 0.65, performing on par with the inter-observer agreement. The dataset and developed methods can facilitate further biomedical research on automated interpretation of lung ultrasound with the potential to expand the clinical utility.
PEP: Parameter Ensembling by Perturbation
Oct 24, 2020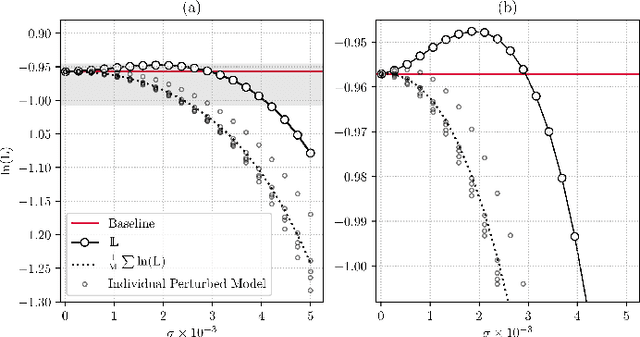
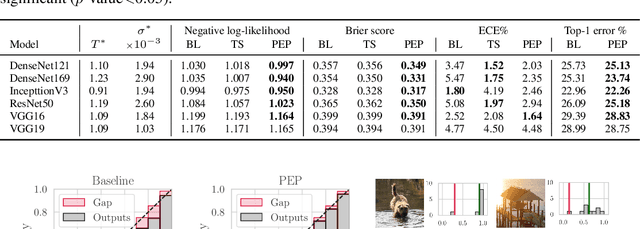
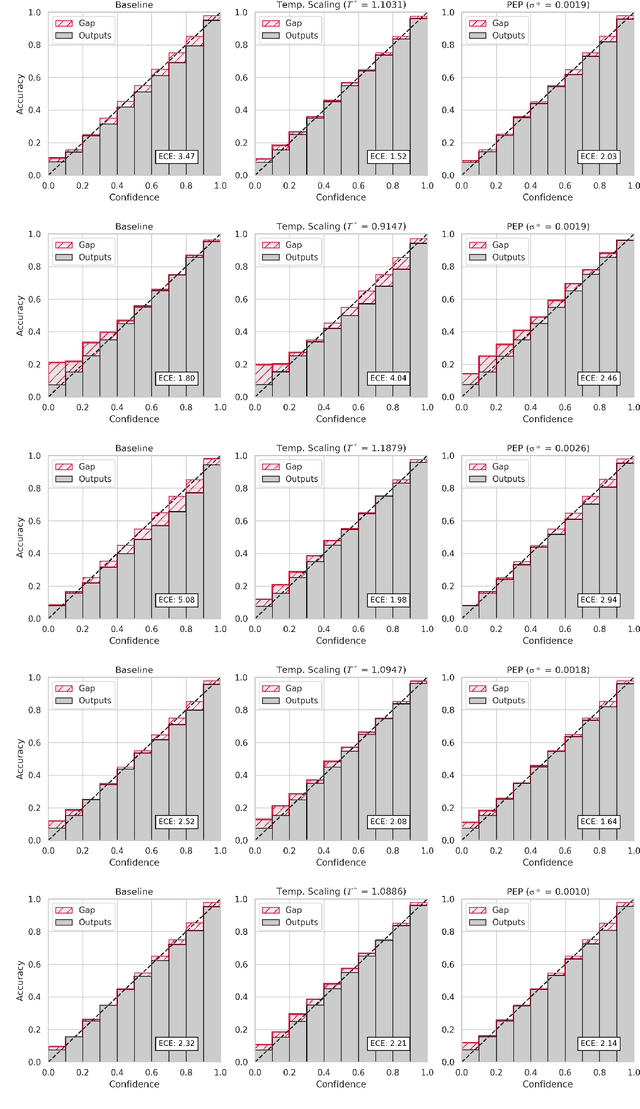
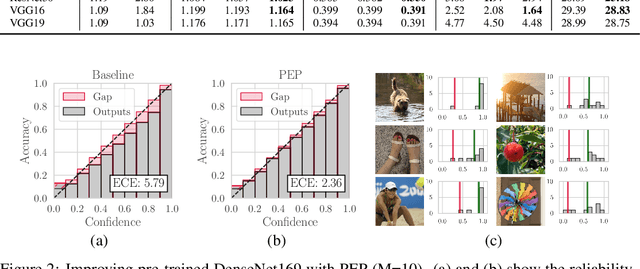
Abstract:Ensembling is now recognized as an effective approach for increasing the predictive performance and calibration of deep networks. We introduce a new approach, Parameter Ensembling by Perturbation (PEP), that constructs an ensemble of parameter values as random perturbations of the optimal parameter set from training by a Gaussian with a single variance parameter. The variance is chosen to maximize the log-likelihood of the ensemble average ($\mathbb{L}$) on the validation data set. Empirically, and perhaps surprisingly, $\mathbb{L}$ has a well-defined maximum as the variance grows from zero (which corresponds to the baseline model). Conveniently, calibration level of predictions also tends to grow favorably until the peak of $\mathbb{L}$ is reached. In most experiments, PEP provides a small improvement in performance, and, in some cases, a substantial improvement in empirical calibration. We show that this "PEP effect" (the gain in log-likelihood) is related to the mean curvature of the likelihood function and the empirical Fisher information. Experiments on ImageNet pre-trained networks including ResNet, DenseNet, and Inception showed improved calibration and likelihood. We further observed a mild improvement in classification accuracy on these networks. Experiments on classification benchmarks such as MNIST and CIFAR-10 showed improved calibration and likelihood, as well as the relationship between the PEP effect and overfitting; this demonstrates that PEP can be used to probe the level of overfitting that occurred during training. In general, no special training procedure or network architecture is needed, and in the case of pre-trained networks, no additional training is needed.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge