Ruben T. Lucassen
Artificial Intelligence-Based Classification of Spitz Tumors
Aug 07, 2025Abstract:Spitz tumors are diagnostically challenging due to overlap in atypical histological features with conventional melanomas. We investigated to what extent AI models, using histological and/or clinical features, can: (1) distinguish Spitz tumors from conventional melanomas; (2) predict the underlying genetic aberration of Spitz tumors; and (3) predict the diagnostic category of Spitz tumors. The AI models were developed and validated using a dataset of 393 Spitz tumors and 379 conventional melanomas. Predictive performance was measured using the AUROC and the accuracy. The performance of the AI models was compared with that of four experienced pathologists in a reader study. Moreover, a simulation experiment was conducted to investigate the impact of implementing AI-based recommendations for ancillary diagnostic testing on the workflow of the pathology department. The best AI model based on UNI features reached an AUROC of 0.95 and an accuracy of 0.86 in differentiating Spitz tumors from conventional melanomas. The genetic aberration was predicted with an accuracy of 0.55 compared to 0.25 for randomly guessing. The diagnostic category was predicted with an accuracy of 0.51, where random chance-level accuracy equaled 0.33. On all three tasks, the AI models performed better than the four pathologists, although differences were not statistically significant for most individual comparisons. Based on the simulation experiment, implementing AI-based recommendations for ancillary diagnostic testing could reduce material costs, turnaround times, and examinations. In conclusion, the AI models achieved a strong predictive performance in distinguishing between Spitz tumors and conventional melanomas. On the more challenging tasks of predicting the genetic aberration and the diagnostic category of Spitz tumors, the AI models performed better than random chance.
Pathology Report Generation and Multimodal Representation Learning for Cutaneous Melanocytic Lesions
Feb 26, 2025Abstract:Millions of melanocytic skin lesions are examined by pathologists each year, the majority of which concern common nevi (i.e., ordinary moles). While most of these lesions can be diagnosed in seconds, writing the corresponding pathology report is much more time-consuming. Automating part of the report writing could, therefore, alleviate the increasing workload of pathologists. In this work, we develop a vision-language model specifically for the pathology domain of cutaneous melanocytic lesions. The model follows the Contrastive Captioner framework and was trained and evaluated using a melanocytic lesion dataset of 42,512 H&E-stained whole slide images and 19,645 corresponding pathology reports. Our results show that the quality scores of model-generated reports were on par with pathologist-written reports for common nevi, assessed by an expert pathologist in a reader study. While report generation revealed to be more difficult for rare melanocytic lesion subtypes, the cross-modal retrieval performance for these cases was considerably better.
On the Importance of Text Preprocessing for Multimodal Representation Learning and Pathology Report Generation
Feb 26, 2025Abstract:Vision-language models in pathology enable multimodal case retrieval and automated report generation. Many of the models developed so far, however, have been trained on pathology reports that include information which cannot be inferred from paired whole slide images (e.g., patient history), potentially leading to hallucinated sentences in generated reports. To this end, we investigate how the selection of information from pathology reports for vision-language modeling affects the quality of the multimodal representations and generated reports. More concretely, we compare a model trained on full reports against a model trained on preprocessed reports that only include sentences describing the cell and tissue appearances based on the H&E-stained slides. For the experiments, we built upon the BLIP-2 framework and used a cutaneous melanocytic lesion dataset of 42,433 H&E-stained whole slide images and 19,636 corresponding pathology reports. Model performance was assessed using image-to-text and text-to-image retrieval, as well as qualitative evaluation of the generated reports by an expert pathologist. Our results demonstrate that text preprocessing prevents hallucination in report generation. Despite the improvement in the quality of the generated reports, training the vision-language model on full reports showed better cross-modal retrieval performance.
Artificial Intelligence-Based Triaging of Cutaneous Melanocytic Lesions
Oct 14, 2024



Abstract:Pathologists are facing an increasing workload due to a growing volume of cases and the need for more comprehensive diagnoses. Aiming to facilitate workload reduction and faster turnaround times, we developed an artificial intelligence (AI) model for triaging cutaneous melanocytic lesions based on whole slide images. The AI model was developed and validated using a retrospective cohort from the UMC Utrecht. The dataset consisted of 52,202 whole slide images from 27,167 unique specimens, acquired from 20,707 patients. Specimens with only common nevi were assigned to the low complexity category (86.6%). In contrast, specimens with any other melanocytic lesion subtype, including non-common nevi, melanocytomas, and melanomas, were assigned to the high complexity category (13.4%). The dataset was split on patient level into a development set (80%) and test sets (20%) for independent evaluation. Predictive performance was primarily measured using the area under the receiver operating characteristic curve (AUROC) and the area under the precision-recall curve (AUPRC). A simulation experiment was performed to study the effect of implementing AI-based triaging in the clinic. The AI model reached an AUROC of 0.966 (95% CI, 0.960-0.972) and an AUPRC of 0.857 (95% CI, 0.836-0.877) on the in-distribution test set, and an AUROC of 0.899 (95% CI, 0.860-0.934) and an AUPRC of 0.498 (95% CI, 0.360-0.639) on the out-of-distribution test set. In the simulation experiment, using random case assignment as baseline, AI-based triaging prevented an average of 43.9 (95% CI, 36-55) initial examinations of high complexity cases by general pathologists for every 500 cases. In conclusion, the AI model achieved a strong predictive performance in differentiating between cutaneous melanocytic lesions of high and low complexity. The improvement in workflow efficiency due to AI-based triaging could be substantial.
Tissue Cross-Section and Pen Marking Segmentation in Whole Slide Images
Jan 24, 2024Abstract:Tissue segmentation is a routine preprocessing step to reduce the computational cost of whole slide image (WSI) analysis by excluding background regions. Traditional image processing techniques are commonly used for tissue segmentation, but often require manual adjustments to parameter values for atypical cases, fail to exclude all slide and scanning artifacts from the background, and are unable to segment adipose tissue. Pen marking artifacts in particular can be a potential source of bias for subsequent analyses if not removed. In addition, several applications require the separation of individual cross-sections, which can be challenging due to tissue fragmentation and adjacent positioning. To address these problems, we develop a convolutional neural network for tissue and pen marking segmentation using a dataset of 200 H&E stained WSIs. For separating tissue cross-sections, we propose a novel post-processing method based on clustering predicted centroid locations of the cross-sections in a 2D histogram. On an independent test set, the model achieved a mean Dice score of 0.981$\pm$0.033 for tissue segmentation and a mean Dice score of 0.912$\pm$0.090 for pen marking segmentation. The mean absolute difference between the number of annotated and separated cross-sections was 0.075$\pm$0.350. Our results demonstrate that the proposed model can accurately segment H&E stained tissue cross-sections and pen markings in WSIs while being robust to many common slide and scanning artifacts. The model with trained model parameters and post-processing method are made publicly available as a Python package called SlideSegmenter.
Deep Learning for Detection and Localization of B-Lines in Lung Ultrasound
Feb 15, 2023



Abstract:Lung ultrasound (LUS) is an important imaging modality used by emergency physicians to assess pulmonary congestion at the patient bedside. B-line artifacts in LUS videos are key findings associated with pulmonary congestion. Not only can the interpretation of LUS be challenging for novice operators, but visual quantification of B-lines remains subject to observer variability. In this work, we investigate the strengths and weaknesses of multiple deep learning approaches for automated B-line detection and localization in LUS videos. We curate and publish, BEDLUS, a new ultrasound dataset comprising 1,419 videos from 113 patients with a total of 15,755 expert-annotated B-lines. Based on this dataset, we present a benchmark of established deep learning methods applied to the task of B-line detection. To pave the way for interpretable quantification of B-lines, we propose a novel "single-point" approach to B-line localization using only the point of origin. Our results show that (a) the area under the receiver operating characteristic curve ranges from 0.864 to 0.955 for the benchmarked detection methods, (b) within this range, the best performance is achieved by models that leverage multiple successive frames as input, and (c) the proposed single-point approach for B-line localization reaches an F1-score of 0.65, performing on par with the inter-observer agreement. The dataset and developed methods can facilitate further biomedical research on automated interpretation of lung ultrasound with the potential to expand the clinical utility.
Corneal Pachymetry by AS-OCT after Descemet's Membrane Endothelial Keratoplasty
Feb 15, 2021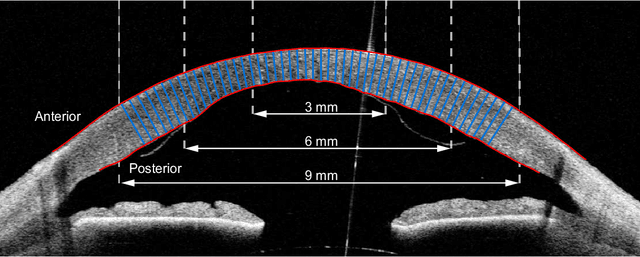
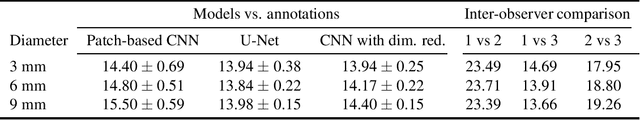
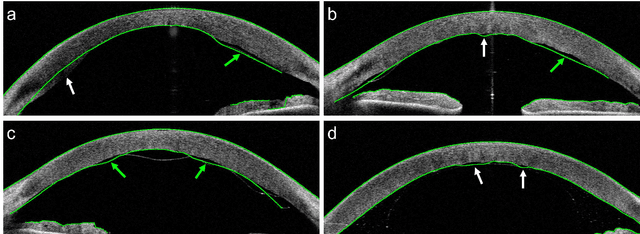
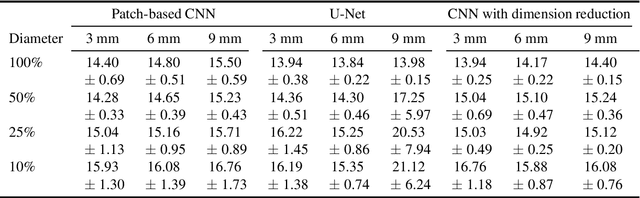
Abstract:Corneal thickness (pachymetry) maps can be used to monitor restoration of corneal endothelial function, for example after Descemet's membrane endothelial keratoplasty (DMEK). Automated delineation of the corneal interfaces in anterior segment optical coherence tomography (AS-OCT) can be challenging for corneas that are irregularly shaped due to pathology, or as a consequence of surgery, leading to incorrect thickness measurements. In this research, deep learning is used to automatically delineate the corneal interfaces and measure corneal thickness with high accuracy in post-DMEK AS-OCT B-scans. Three different deep learning strategies were developed based on 960 B-scans from 68 patients. On an independent test set of 320 B-scans, corneal thickness could be measured with an error of 13.98 to 15.50 micrometer for the central 9 mm range, which is less than 3% of the average corneal thickness. The accurate thickness measurements were used to construct detailed pachymetry maps. Moreover, follow-up scans could be registered based on anatomical landmarks to obtain differential pachymetry maps. These maps may enable a more comprehensive understanding of the restoration of the endothelial function after DMEK, where thickness often varies throughout different regions of the cornea, and subsequently contribute to a standardized postoperative regime.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge