Mohammad H. Jafari
Deep Learning for Detection and Localization of B-Lines in Lung Ultrasound
Feb 15, 2023



Abstract:Lung ultrasound (LUS) is an important imaging modality used by emergency physicians to assess pulmonary congestion at the patient bedside. B-line artifacts in LUS videos are key findings associated with pulmonary congestion. Not only can the interpretation of LUS be challenging for novice operators, but visual quantification of B-lines remains subject to observer variability. In this work, we investigate the strengths and weaknesses of multiple deep learning approaches for automated B-line detection and localization in LUS videos. We curate and publish, BEDLUS, a new ultrasound dataset comprising 1,419 videos from 113 patients with a total of 15,755 expert-annotated B-lines. Based on this dataset, we present a benchmark of established deep learning methods applied to the task of B-line detection. To pave the way for interpretable quantification of B-lines, we propose a novel "single-point" approach to B-line localization using only the point of origin. Our results show that (a) the area under the receiver operating characteristic curve ranges from 0.864 to 0.955 for the benchmarked detection methods, (b) within this range, the best performance is achieved by models that leverage multiple successive frames as input, and (c) the proposed single-point approach for B-line localization reaches an F1-score of 0.65, performing on par with the inter-observer agreement. The dataset and developed methods can facilitate further biomedical research on automated interpretation of lung ultrasound with the potential to expand the clinical utility.
U-LanD: Uncertainty-Driven Video Landmark Detection
Feb 02, 2021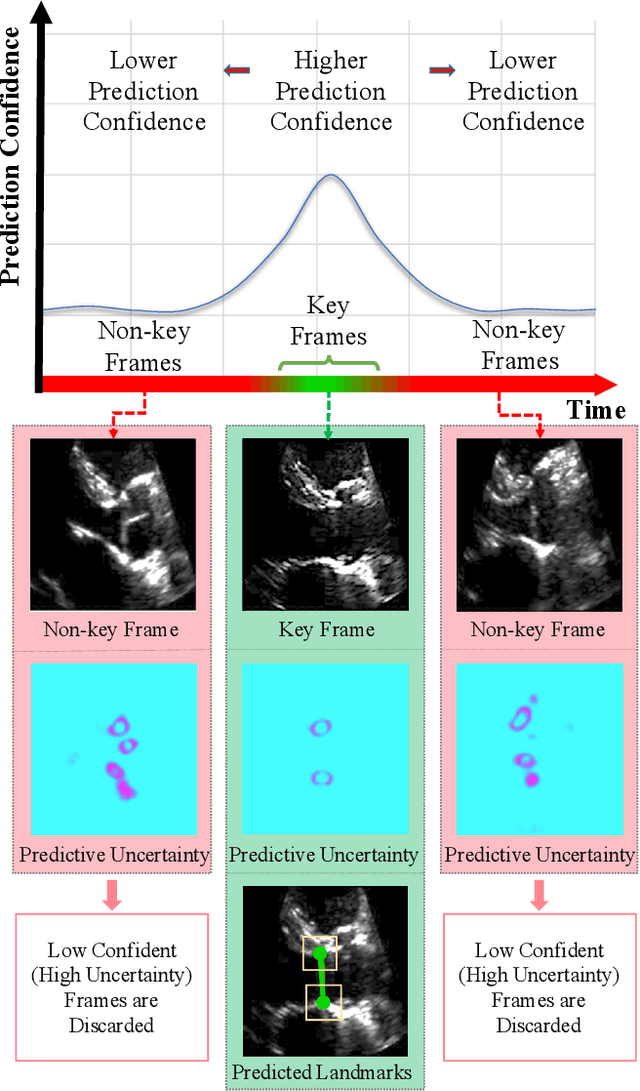
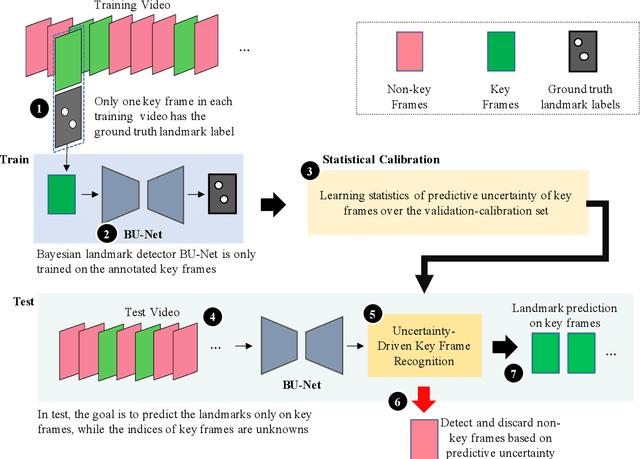
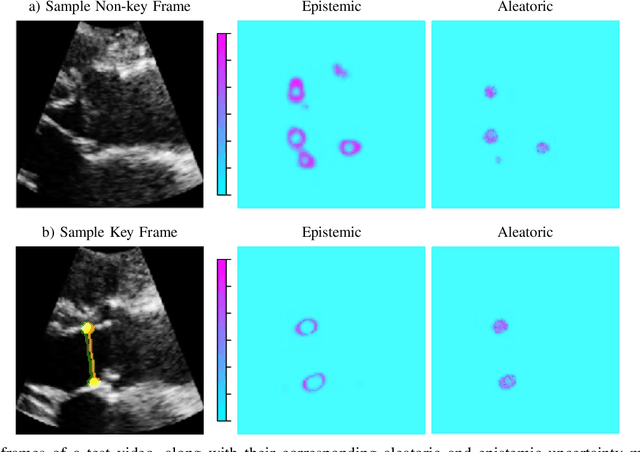
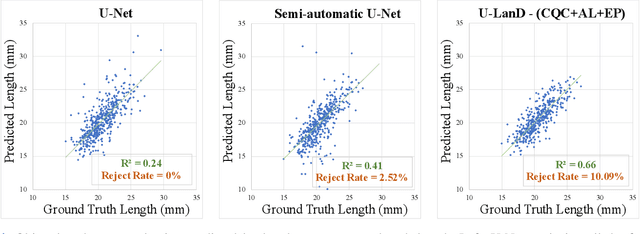
Abstract:This paper presents U-LanD, a framework for joint detection of key frames and landmarks in videos. We tackle a specifically challenging problem, where training labels are noisy and highly sparse. U-LanD builds upon a pivotal observation: a deep Bayesian landmark detector solely trained on key video frames, has significantly lower predictive uncertainty on those frames vs. other frames in videos. We use this observation as an unsupervised signal to automatically recognize key frames on which we detect landmarks. As a test-bed for our framework, we use ultrasound imaging videos of the heart, where sparse and noisy clinical labels are only available for a single frame in each video. Using data from 4,493 patients, we demonstrate that U-LanD can exceedingly outperform the state-of-the-art non-Bayesian counterpart by a noticeable absolute margin of 42% in R2 score, with almost no overhead imposed on the model size. Our approach is generic and can be potentially applied to other challenging data with noisy and sparse training labels.
A Study into Echocardiography View Conversion
Dec 05, 2019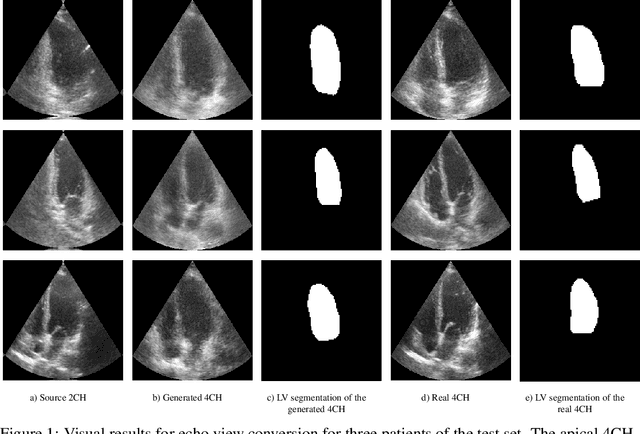
Abstract:Transthoracic echo is one of the most common means of cardiac studies in the clinical routines. During the echo exam, the sonographer captures a set of standard cross sections (echo views) of the heart. Each 2D echo view cuts through the 3D cardiac geometry via a unique plane. Consequently, different views share some limited information. In this work, we investigate the feasibility of generating a 2D echo view using another view based on adversarial generative models. The objective optimized to train the view-conversion model is based on the ideas introduced by LSGAN, PatchGAN and Conditional GAN (cGAN). The size and length of the left ventricle in the generated target echo view is compared against that of the target ground-truth to assess the validity of the echo view conversion. Results show that there is a correlation of 0.50 between the LV areas and 0.49 between the LV lengths of the generated target frames and the real target frames.
Dense Fully Convolutional Network for Skin Lesion Segmentation
Jun 05, 2018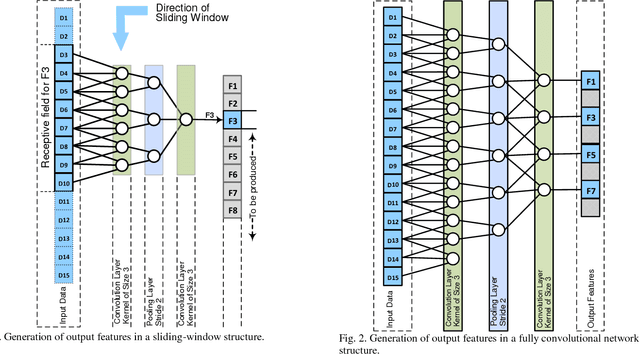

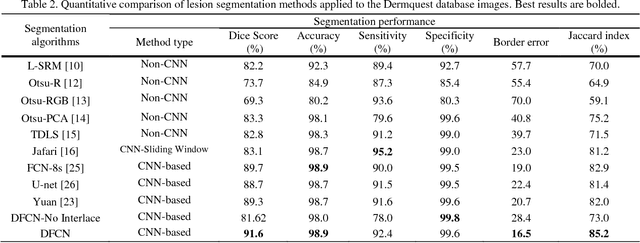
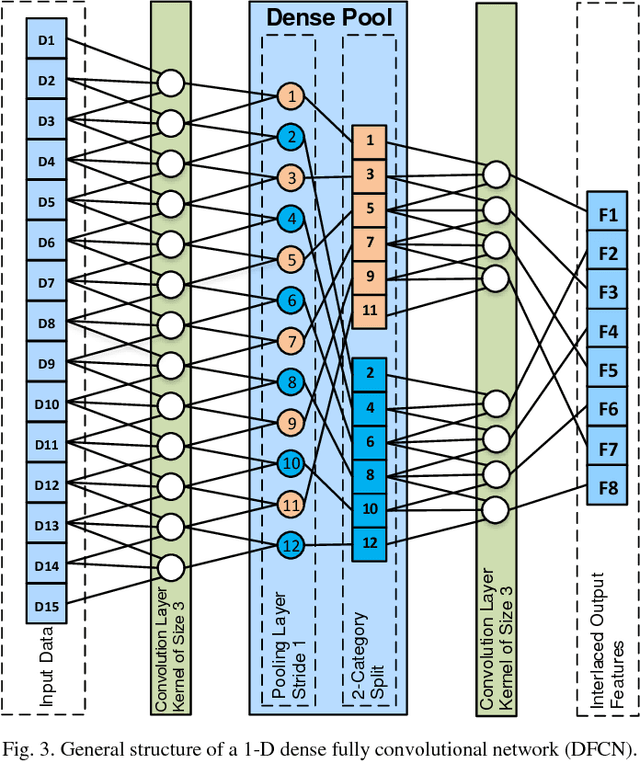
Abstract:Lesion segmentation in skin images is an important step in computerized detection of skin cancer. Melanoma is known as one of the most life threatening types of this cancer. Existing methods often fall short of accurately segmenting lesions with fuzzy boarders. In this paper, a new class of fully convolutional network is proposed, with new dense pooling layers for segmentation of lesion regions in non-dermoscopic images. Unlike other existing convolutional networks, this proposed network is designed to produce dense feature maps. This network leads to highly accurate segmentation of lesions. The produced dice score here is 91.6% which outperforms state-of-the-art algorithms in segmentation of skin lesions based on the Dermquest dataset.
Extraction of Skin Lesions from Non-Dermoscopic Images Using Deep Learning
Sep 08, 2016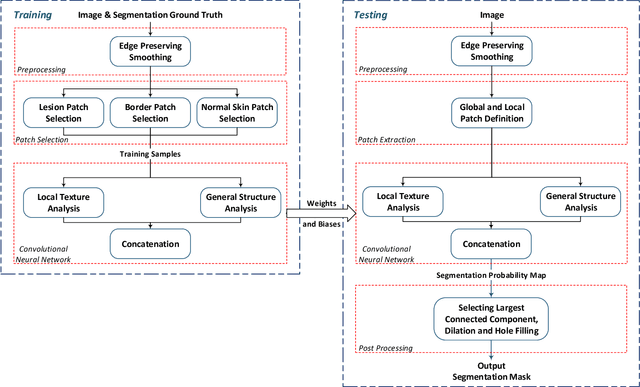
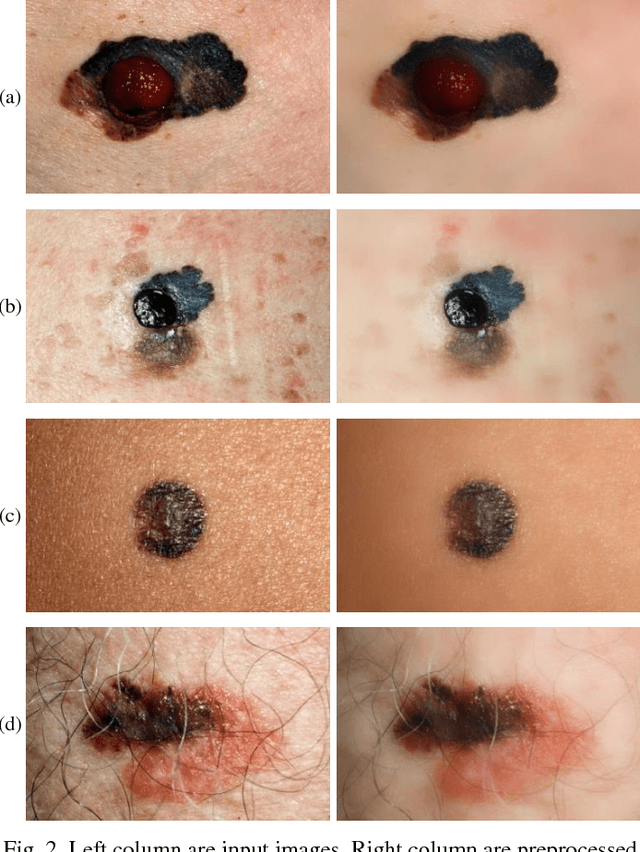
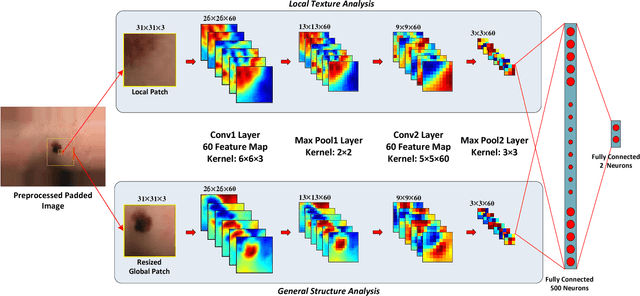
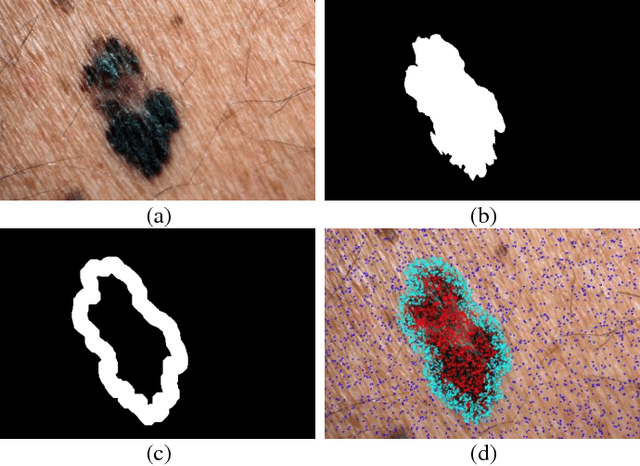
Abstract:Melanoma is amongst most aggressive types of cancer. However, it is highly curable if detected in its early stages. Prescreening of suspicious moles and lesions for malignancy is of great importance. Detection can be done by images captured by standard cameras, which are more preferable due to low cost and availability. One important step in computerized evaluation of skin lesions is accurate detection of lesion region, i.e. segmentation of an image into two regions as lesion and normal skin. Accurate segmentation can be challenging due to burdens such as illumination variation and low contrast between lesion and healthy skin. In this paper, a method based on deep neural networks is proposed for accurate extraction of a lesion region. The input image is preprocessed and then its patches are fed to a convolutional neural network (CNN). Local texture and global structure of the patches are processed in order to assign pixels to lesion or normal classes. A method for effective selection of training patches is used for more accurate detection of a lesion border. The output segmentation mask is refined by some post processing operations. The experimental results of qualitative and quantitative evaluations demonstrate that our method can outperform other state-of-the-art algorithms exist in the literature.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge