Ang Nan Gu
Reliable Multi-View Learning with Conformal Prediction for Aortic Stenosis Classification in Echocardiography
Sep 15, 2024Abstract:The fundamental problem with ultrasound-guided diagnosis is that the acquired images are often 2-D cross-sections of a 3-D anatomy, potentially missing important anatomical details. This limitation leads to challenges in ultrasound echocardiography, such as poor visualization of heart valves or foreshortening of ventricles. Clinicians must interpret these images with inherent uncertainty, a nuance absent in machine learning's one-hot labels. We propose Re-Training for Uncertainty (RT4U), a data-centric method to introduce uncertainty to weakly informative inputs in the training set. This simple approach can be incorporated to existing state-of-the-art aortic stenosis classification methods to further improve their accuracy. When combined with conformal prediction techniques, RT4U can yield adaptively sized prediction sets which are guaranteed to contain the ground truth class to a high accuracy. We validate the effectiveness of RT4U on three diverse datasets: a public (TMED-2) and a private AS dataset, along with a CIFAR-10-derived toy dataset. Results show improvement on all the datasets.
ProtoASNet: Dynamic Prototypes for Inherently Interpretable and Uncertainty-Aware Aortic Stenosis Classification in Echocardiography
Jul 26, 2023


Abstract:Aortic stenosis (AS) is a common heart valve disease that requires accurate and timely diagnosis for appropriate treatment. Most current automatic AS severity detection methods rely on black-box models with a low level of trustworthiness, which hinders clinical adoption. To address this issue, we propose ProtoASNet, a prototypical network that directly detects AS from B-mode echocardiography videos, while making interpretable predictions based on the similarity between the input and learned spatio-temporal prototypes. This approach provides supporting evidence that is clinically relevant, as the prototypes typically highlight markers such as calcification and restricted movement of aortic valve leaflets. Moreover, ProtoASNet utilizes abstention loss to estimate aleatoric uncertainty by defining a set of prototypes that capture ambiguity and insufficient information in the observed data. This provides a reliable system that can detect and explain when it may fail. We evaluate ProtoASNet on a private dataset and the publicly available TMED-2 dataset, where it outperforms existing state-of-the-art methods with an accuracy of 80.0% and 79.7%, respectively. Furthermore, ProtoASNet provides interpretability and an uncertainty measure for each prediction, which can improve transparency and facilitate the interactive usage of deep networks to aid clinical decision-making. Our source code is available at: https://github.com/hooman007/ProtoASNet.
U-LanD: Uncertainty-Driven Video Landmark Detection
Feb 02, 2021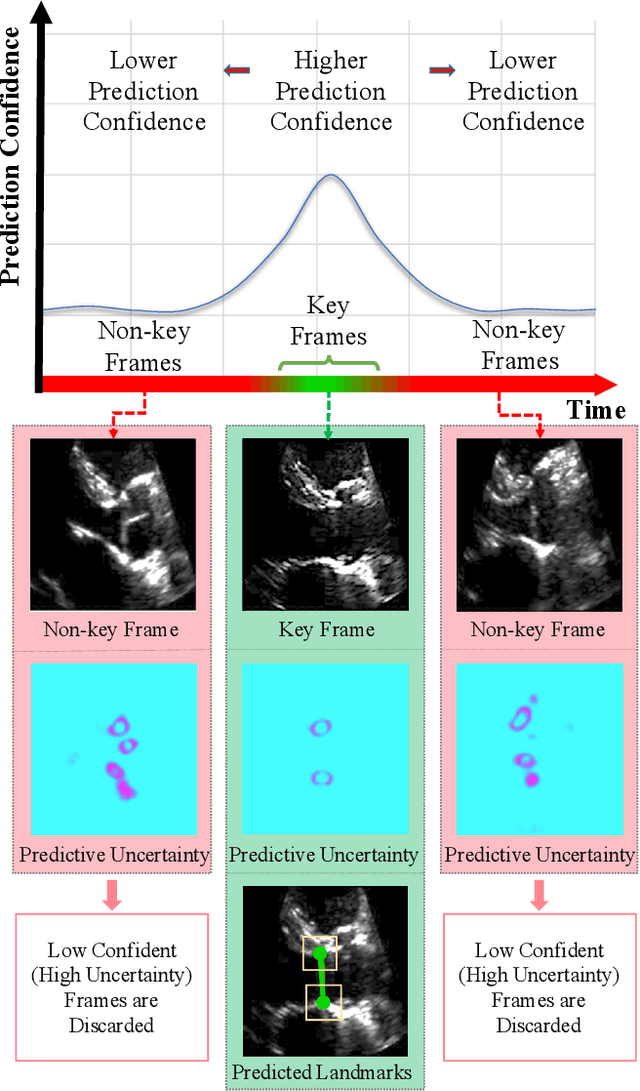
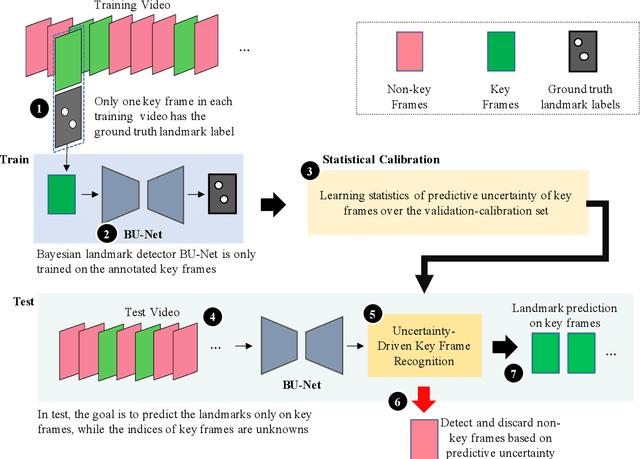
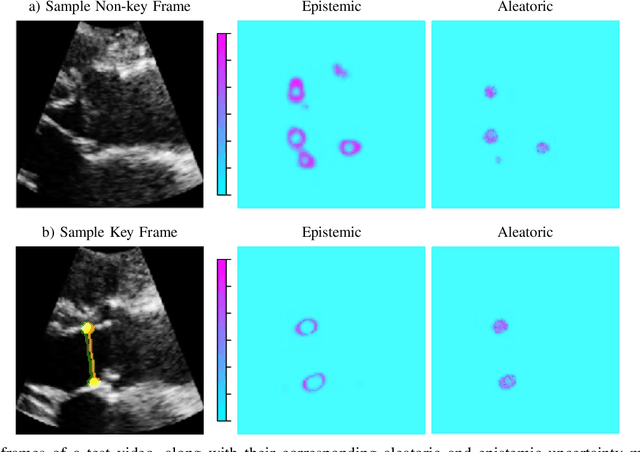
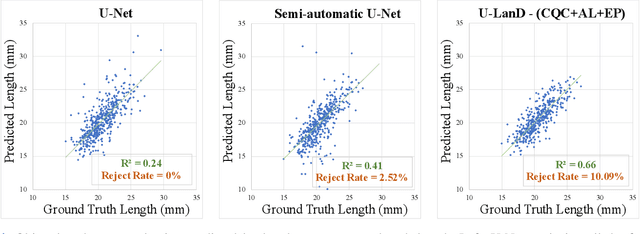
Abstract:This paper presents U-LanD, a framework for joint detection of key frames and landmarks in videos. We tackle a specifically challenging problem, where training labels are noisy and highly sparse. U-LanD builds upon a pivotal observation: a deep Bayesian landmark detector solely trained on key video frames, has significantly lower predictive uncertainty on those frames vs. other frames in videos. We use this observation as an unsupervised signal to automatically recognize key frames on which we detect landmarks. As a test-bed for our framework, we use ultrasound imaging videos of the heart, where sparse and noisy clinical labels are only available for a single frame in each video. Using data from 4,493 patients, we demonstrate that U-LanD can exceedingly outperform the state-of-the-art non-Bayesian counterpart by a noticeable absolute margin of 42% in R2 score, with almost no overhead imposed on the model size. Our approach is generic and can be potentially applied to other challenging data with noisy and sparse training labels.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge