Armin Saadat
Point Tracking as a Temporal Cue for Robust Myocardial Segmentation in Echocardiography Videos
Jan 14, 2026Abstract:Purpose: Myocardium segmentation in echocardiography videos is a challenging task due to low contrast, noise, and anatomical variability. Traditional deep learning models either process frames independently, ignoring temporal information, or rely on memory-based feature propagation, which accumulates error over time. Methods: We propose Point-Seg, a transformer-based segmentation framework that integrates point tracking as a temporal cue to ensure stable and consistent segmentation of myocardium across frames. Our method leverages a point-tracking module trained on a synthetic echocardiography dataset to track key anatomical landmarks across video sequences. These tracked trajectories provide an explicit motion-aware signal that guides segmentation, reducing drift and eliminating the need for memory-based feature accumulation. Additionally, we incorporate a temporal smoothing loss to further enhance temporal consistency across frames. Results: We evaluate our approach on both public and private echocardiography datasets. Experimental results demonstrate that Point-Seg has statistically similar accuracy in terms of Dice to state-of-the-art segmentation models in high quality echo data, while it achieves better segmentation accuracy in lower quality echo with improved temporal stability. Furthermore, Point-Seg has the key advantage of pixel-level myocardium motion information as opposed to other segmentation methods. Such information is essential in the computation of other downstream tasks such as myocardial strain measurement and regional wall motion abnormality detection. Conclusion: Point-Seg demonstrates that point tracking can serve as an effective temporal cue for consistent video segmentation, offering a reliable and generalizable approach for myocardium segmentation in echocardiography videos. The code is available at https://github.com/DeepRCL/PointSeg.
Federated Impression for Learning with Distributed Heterogeneous Data
Sep 11, 2024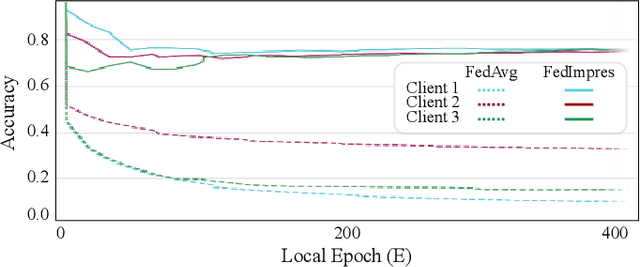
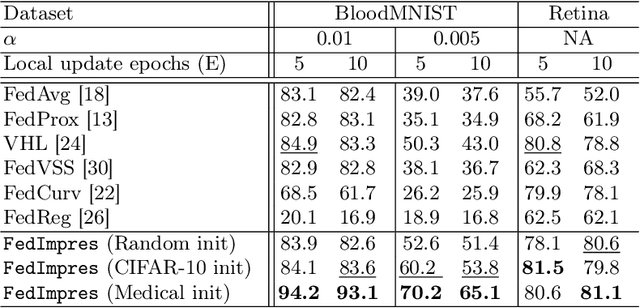
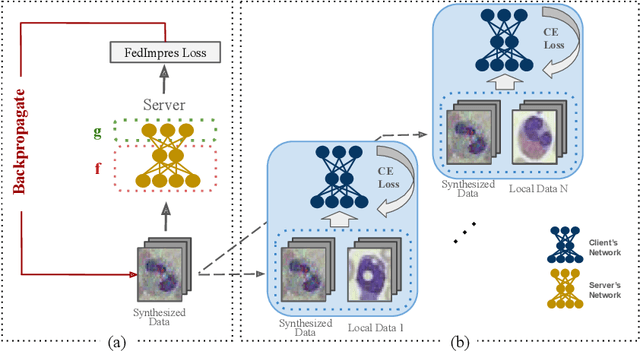
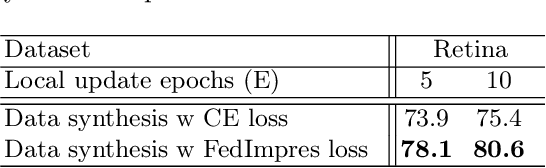
Abstract:Standard deep learning-based classification approaches may not always be practical in real-world clinical applications, as they require a centralized collection of all samples. Federated learning (FL) provides a paradigm that can learn from distributed datasets across clients without requiring them to share data, which can help mitigate privacy and data ownership issues. In FL, sub-optimal convergence caused by data heterogeneity is common among data from different health centers due to the variety in data collection protocols and patient demographics across centers. Through experimentation in this study, we show that data heterogeneity leads to the phenomenon of catastrophic forgetting during local training. We propose FedImpres which alleviates catastrophic forgetting by restoring synthetic data that represents the global information as federated impression. To achieve this, we distill the global model resulting from each communication round. Subsequently, we use the synthetic data alongside the local data to enhance the generalization of local training. Extensive experiments show that the proposed method achieves state-of-the-art performance on both the BloodMNIST and Retina datasets, which contain label imbalance and domain shift, with an improvement in classification accuracy of up to 20%.
ProtoASNet: Dynamic Prototypes for Inherently Interpretable and Uncertainty-Aware Aortic Stenosis Classification in Echocardiography
Jul 26, 2023


Abstract:Aortic stenosis (AS) is a common heart valve disease that requires accurate and timely diagnosis for appropriate treatment. Most current automatic AS severity detection methods rely on black-box models with a low level of trustworthiness, which hinders clinical adoption. To address this issue, we propose ProtoASNet, a prototypical network that directly detects AS from B-mode echocardiography videos, while making interpretable predictions based on the similarity between the input and learned spatio-temporal prototypes. This approach provides supporting evidence that is clinically relevant, as the prototypes typically highlight markers such as calcification and restricted movement of aortic valve leaflets. Moreover, ProtoASNet utilizes abstention loss to estimate aleatoric uncertainty by defining a set of prototypes that capture ambiguity and insufficient information in the observed data. This provides a reliable system that can detect and explain when it may fail. We evaluate ProtoASNet on a private dataset and the publicly available TMED-2 dataset, where it outperforms existing state-of-the-art methods with an accuracy of 80.0% and 79.7%, respectively. Furthermore, ProtoASNet provides interpretability and an uncertainty measure for each prediction, which can improve transparency and facilitate the interactive usage of deep networks to aid clinical decision-making. Our source code is available at: https://github.com/hooman007/ProtoASNet.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge