Sana Ayromlou
Adaptive Latent-Space Constraints in Personalized FL
May 12, 2025Abstract:Federated learning (FL) has become an effective and widely used approach to training deep learning models on decentralized datasets held by distinct clients. FL also strengthens both security and privacy protections for training data. Common challenges associated with statistical heterogeneity between distributed datasets have spurred significant interest in personalized FL (pFL) methods, where models combine aspects of global learning with local modeling specific to each client's unique characteristics. In this work, the efficacy of theoretically supported, adaptive MMD measures within the Ditto framework, a state-of-the-art technique in pFL, are investigated. The use of such measures significantly improves model performance across a variety of tasks, especially those with pronounced feature heterogeneity. While the Ditto algorithm is specifically considered, such measures are directly applicable to a number of other pFL settings, and the results motivate the use of constraints tailored to the various kinds of heterogeneity expected in FL systems.
Federated Impression for Learning with Distributed Heterogeneous Data
Sep 11, 2024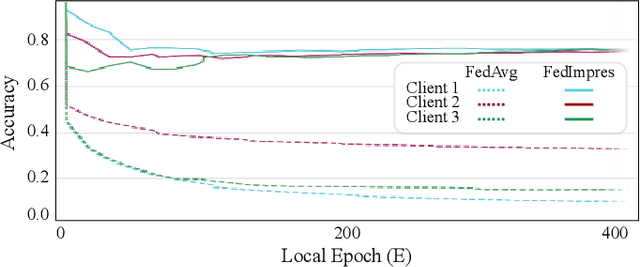
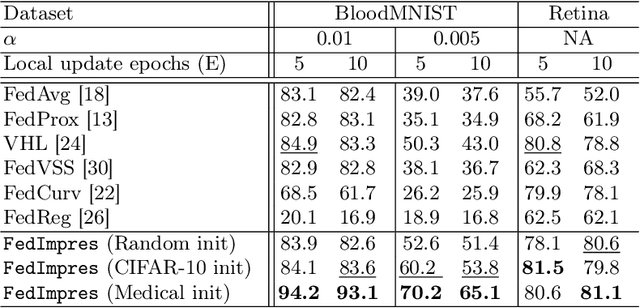
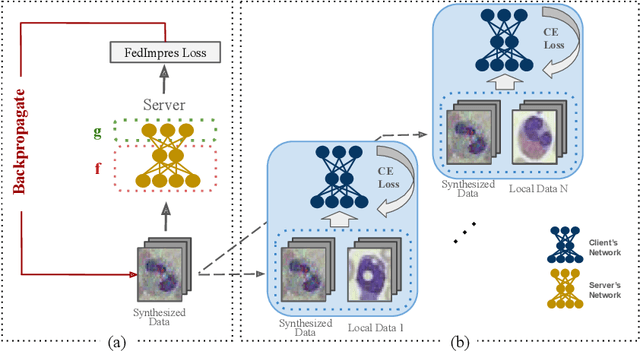
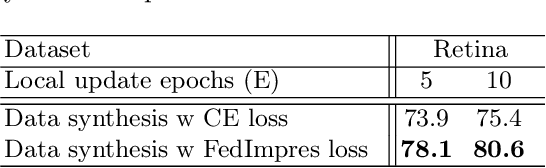
Abstract:Standard deep learning-based classification approaches may not always be practical in real-world clinical applications, as they require a centralized collection of all samples. Federated learning (FL) provides a paradigm that can learn from distributed datasets across clients without requiring them to share data, which can help mitigate privacy and data ownership issues. In FL, sub-optimal convergence caused by data heterogeneity is common among data from different health centers due to the variety in data collection protocols and patient demographics across centers. Through experimentation in this study, we show that data heterogeneity leads to the phenomenon of catastrophic forgetting during local training. We propose FedImpres which alleviates catastrophic forgetting by restoring synthetic data that represents the global information as federated impression. To achieve this, we distill the global model resulting from each communication round. Subsequently, we use the synthetic data alongside the local data to enhance the generalization of local training. Extensive experiments show that the proposed method achieves state-of-the-art performance on both the BloodMNIST and Retina datasets, which contain label imbalance and domain shift, with an improvement in classification accuracy of up to 20%.
CCSI: Continual Class-Specific Impression for Data-free Class Incremental Learning
Jun 09, 2024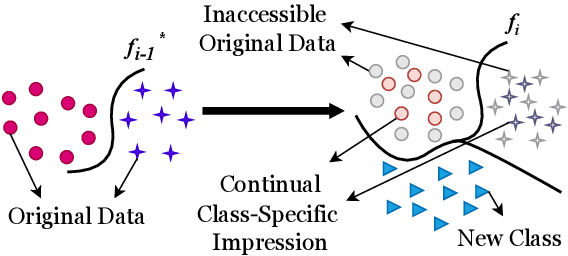

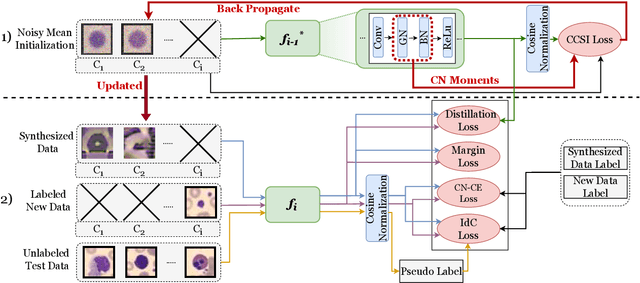
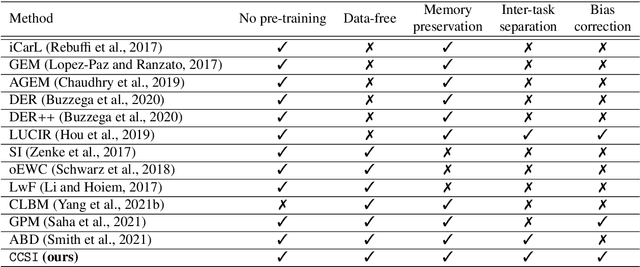
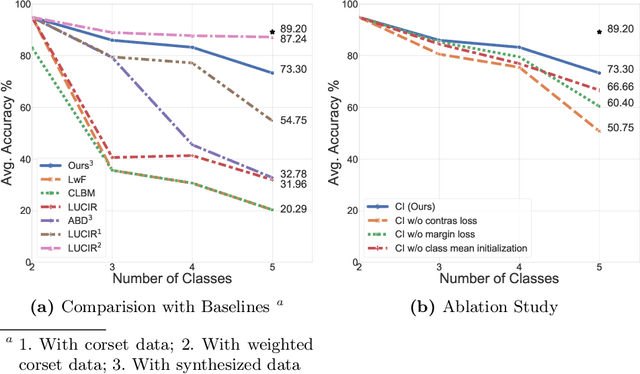
Abstract:In real-world clinical settings, traditional deep learning-based classification methods struggle with diagnosing newly introduced disease types because they require samples from all disease classes for offline training. Class incremental learning offers a promising solution by adapting a deep network trained on specific disease classes to handle new diseases. However, catastrophic forgetting occurs, decreasing the performance of earlier classes when adapting the model to new data. Prior proposed methodologies to overcome this require perpetual storage of previous samples, posing potential practical concerns regarding privacy and storage regulations in healthcare. To this end, we propose a novel data-free class incremental learning framework that utilizes data synthesis on learned classes instead of data storage from previous classes. Our key contributions include acquiring synthetic data known as Continual Class-Specific Impression (CCSI) for previously inaccessible trained classes and presenting a methodology to effectively utilize this data for updating networks when introducing new classes. We obtain CCSI by employing data inversion over gradients of the trained classification model on previous classes starting from the mean image of each class inspired by common landmarks shared among medical images and utilizing continual normalization layers statistics as a regularizer in this pixel-wise optimization process. Subsequently, we update the network by combining the synthesized data with new class data and incorporate several losses, including an intra-domain contrastive loss to generalize the deep network trained on the synthesized data to real data, a margin loss to increase separation among previous classes and new ones, and a cosine-normalized cross-entropy loss to alleviate the adverse effects of imbalanced distributions in training data.
Can Generative Models Improve Self-Supervised Representation Learning?
Mar 09, 2024



Abstract:The rapid advancement in self-supervised learning (SSL) has highlighted its potential to leverage unlabeled data for learning powerful visual representations. However, existing SSL approaches, particularly those employing different views of the same image, often rely on a limited set of predefined data augmentations. This constrains the diversity and quality of transformations, which leads to sub-optimal representations. In this paper, we introduce a novel framework that enriches the SSL paradigm by utilizing generative models to produce semantically consistent image augmentations. By directly conditioning generative models on a source image representation, our method enables the generation of diverse augmentations while maintaining the semantics of the source image, thus offering a richer set of data for self-supervised learning. Our experimental results demonstrate that our framework significantly enhances the quality of learned visual representations. This research demonstrates that incorporating generative models into the SSL workflow opens new avenues for exploring the potential of unlabeled visual data. This development paves the way for more robust and versatile representation learning techniques.
Class Impression for Data-free Incremental Learning
Jul 04, 2022
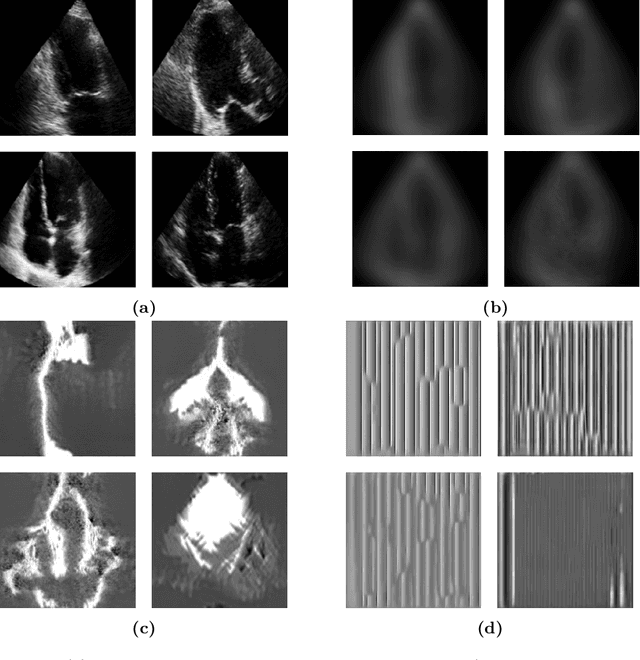
Abstract:Standard deep learning-based classification approaches require collecting all samples from all classes in advance and are trained offline. This paradigm may not be practical in real-world clinical applications, where new classes are incrementally introduced through the addition of new data. Class incremental learning is a strategy allowing learning from such data. However, a major challenge is catastrophic forgetting, i.e., performance degradation on previous classes when adapting a trained model to new data. Prior methodologies to alleviate this challenge save a portion of training data require perpetual storage of such data that may introduce privacy issues. Here, we propose a novel data-free class incremental learning framework that first synthesizes data from the model trained on previous classes to generate a \ours. Subsequently, it updates the model by combining the synthesized data with new class data. Furthermore, we incorporate a cosine normalized Cross-entropy loss to mitigate the adverse effects of the imbalance, a margin loss to increase separation among previous classes and new ones, and an intra-domain contrastive loss to generalize the model trained on the synthesized data to real data. We compare our proposed framework with state-of-the-art methods in class incremental learning, where we demonstrate improvement in accuracy for the classification of 11,062 echocardiography cine series of patients.
SVG-Net: An SVG-based Trajectory Prediction Model
Oct 11, 2021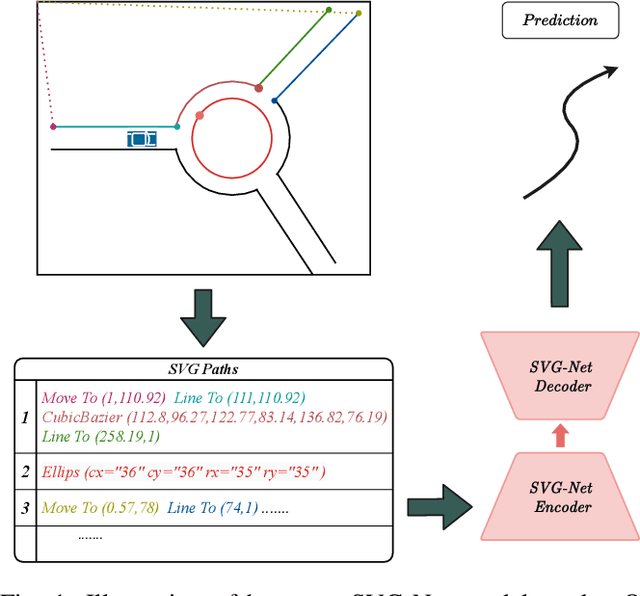
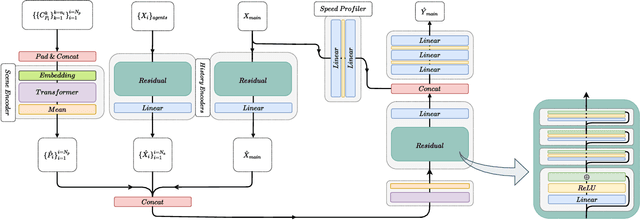
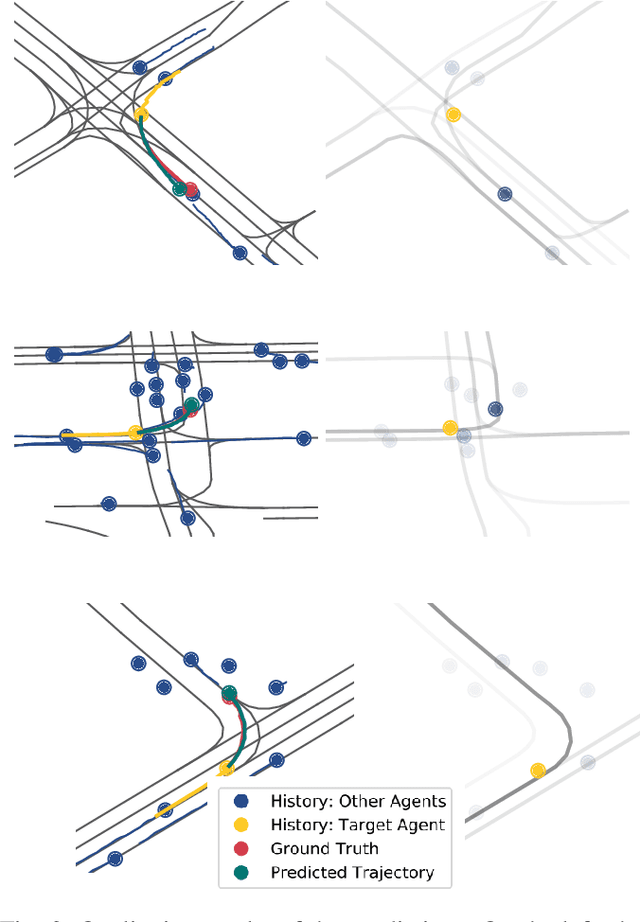
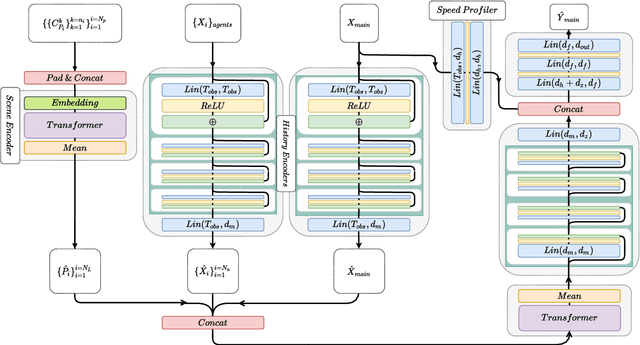
Abstract:Anticipating motions of vehicles in a scene is an essential problem for safe autonomous driving systems. To this end, the comprehension of the scene's infrastructure is often the main clue for predicting future trajectories. Most of the proposed approaches represent the scene with a rasterized format and some of the more recent approaches leverage custom vectorized formats. In contrast, we propose representing the scene's information by employing Scalable Vector Graphics (SVG). SVG is a well-established format that matches the problem of trajectory prediction better than rasterized formats while being more general than arbitrary vectorized formats. SVG has the potential to provide the convenience and generality of raster-based solutions if coupled with a powerful tool such as CNNs, for which we introduce SVG-Net. SVG-Net is a Transformer-based Neural Network that can effectively capture the scene's information from SVG inputs. Thanks to the self-attention mechanism in its Transformers, SVG-Net can also adequately apprehend relations amongst the scene and the agents. We demonstrate SVG-Net's effectiveness by evaluating its performance on the publicly available Argoverse forecasting dataset. Finally, we illustrate how, by using SVG, one can benefit from datasets and advancements in other research fronts that also utilize the same input format. Our code is available at https://vita-epfl.github.io/SVGNet/.
Dementia Severity Classification under Small Sample Size and Weak Supervision in Thick Slice MRI
Mar 18, 2021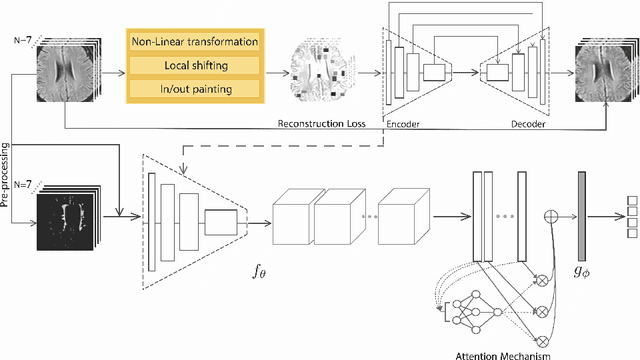
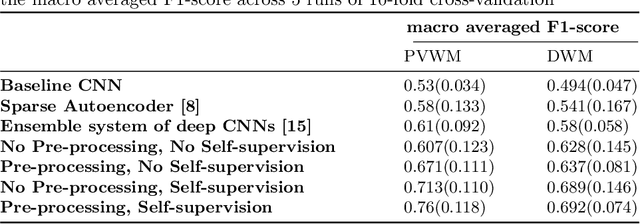
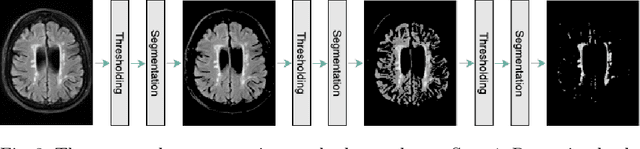
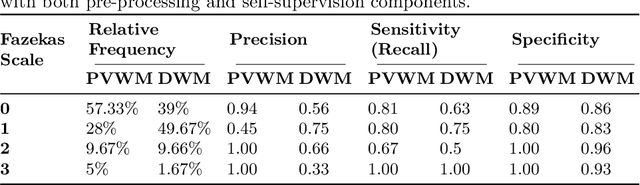
Abstract:Early detection of dementia through specific biomarkers in MR images plays a critical role in developing support strategies proactively. Fazekas scale facilitates an accurate quantitative assessment of the severity of white matter lesions and hence the disease. Imaging Biomarkers of dementia are multiple and comprehensive documentation of them is time-consuming. Therefore, any effort to automatically extract these biomarkers will be of clinical value while reducing inter-rater discrepancies. To tackle this problem, we propose to classify the disease severity based on the Fazekas scale through the visual biomarkers, namely the Periventricular White Matter (PVWM) and the Deep White Matter (DWM) changes, in the real-world setting of thick-slice MRI. Small training sample size and weak supervision in form of assigning severity labels to the whole MRI stack are among the main challenges. To combat the mentioned issues, we have developed a deep learning pipeline that employs self-supervised representation learning, multiple instance learning, and appropriate pre-processing steps. We use pretext tasks such as non-linear transformation, local shuffling, in- and out-painting for self-supervised learning of useful features in this domain. Furthermore, an attention model is used to determine the relevance of each MRI slice for predicting the Fazekas scale in an unsupervised manner. We show the significant superiority of our method in distinguishing different classes of dementia compared to state-of-the-art methods in our mentioned setting, which improves the macro averaged F1-score of state-of-the-art from 61% to 76% in PVWM, and from 58% to 69.2% in DWM.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge