Hamid R. Rabiee
AI-Med Group, AI Innovation Center, Sharif University of Technology, Tehran, Iran, DML Lab, Department of Computer Engineering, Sharif University of Technology, Tehran, Iran
ReDiF: Reinforced Distillation for Few Step Diffusion
Dec 28, 2025Abstract:Distillation addresses the slow sampling problem in diffusion models by creating models with smaller size or fewer steps that approximate the behavior of high-step teachers. In this work, we propose a reinforcement learning based distillation framework for diffusion models. Instead of relying on fixed reconstruction or consistency losses, we treat the distillation process as a policy optimization problem, where the student is trained using a reward signal derived from alignment with the teacher's outputs. This RL driven approach dynamically guides the student to explore multiple denoising paths, allowing it to take longer, optimized steps toward high-probability regions of the data distribution, rather than relying on incremental refinements. Our framework utilizes the inherent ability of diffusion models to handle larger steps and effectively manage the generative process. Experimental results show that our method achieves superior performance with significantly fewer inference steps and computational resources compared to existing distillation techniques. Additionally, the framework is model agnostic, applicable to any type of diffusion models with suitable reward functions, providing a general optimization paradigm for efficient diffusion learning.
RealDrag: The First Dragging Benchmark with Real Target Image
Dec 13, 2025Abstract:The evaluation of drag based image editing models is unreliable due to a lack of standardized benchmarks and metrics. This ambiguity stems from inconsistent evaluation protocols and, critically, the absence of datasets containing ground truth target images, making objective comparisons between competing methods difficult. To address this, we introduce \textbf{RealDrag}, the first comprehensive benchmark for point based image editing that includes paired ground truth target images. Our dataset contains over 400 human annotated samples from diverse video sources, providing source/target images, handle/target points, editable region masks, and descriptive captions for both the image and the editing action. We also propose four novel, task specific metrics: Semantical Distance (SeD), Outer Mask Preserving Score (OMPS), Inner Patch Preserving Score (IPPS), and Directional Similarity (DiS). These metrics are designed to quantify pixel level matching fidelity, check preservation of non edited (out of mask) regions, and measure semantic alignment with the desired task. Using this benchmark, we conduct the first large scale systematic analysis of the field, evaluating 17 SOTA models. Our results reveal clear trade offs among current approaches and establish a robust, reproducible baseline to guide future research. Our dataset and evaluation toolkit will be made publicly available.
CineLOG: A Training Free Approach for Cinematic Long Video Generation
Dec 13, 2025Abstract:Controllable video synthesis is a central challenge in computer vision, yet current models struggle with fine grained control beyond textual prompts, particularly for cinematic attributes like camera trajectory and genre. Existing datasets often suffer from severe data imbalance, noisy labels, or a significant simulation to real gap. To address this, we introduce CineLOG, a new dataset of 5,000 high quality, balanced, and uncut video clips. Each entry is annotated with a detailed scene description, explicit camera instructions based on a standard cinematic taxonomy, and genre label, ensuring balanced coverage across 17 diverse camera movements and 15 film genres. We also present our novel pipeline designed to create this dataset, which decouples the complex text to video (T2V) generation task into four easier stages with more mature technology. To enable coherent, multi shot sequences, we introduce a novel Trajectory Guided Transition Module that generates smooth spatio-temporal interpolation. Extensive human evaluations show that our pipeline significantly outperforms SOTA end to end T2V models in adhering to specific camera and screenplay instructions, while maintaining professional visual quality. All codes and data are available at https://cine-log.pages.dev.
Beyond Unified Models: A Service-Oriented Approach to Low Latency, Context Aware Phonemization for Real Time TTS
Dec 08, 2025Abstract:Lightweight, real-time text-to-speech systems are crucial for accessibility. However, the most efficient TTS models often rely on lightweight phonemizers that struggle with context-dependent challenges. In contrast, more advanced phonemizers with a deeper linguistic understanding typically incur high computational costs, which prevents real-time performance. This paper examines the trade-off between phonemization quality and inference speed in G2P-aided TTS systems, introducing a practical framework to bridge this gap. We propose lightweight strategies for context-aware phonemization and a service-oriented TTS architecture that executes these modules as independent services. This design decouples heavy context-aware components from the core TTS engine, effectively breaking the latency barrier and enabling real-time use of high-quality phonemization models. Experimental results confirm that the proposed system improves pronunciation soundness and linguistic accuracy while maintaining real-time responsiveness, making it well-suited for offline and end-device TTS applications.
FIRE: Faithful Interpretable Recommendation Explanations
Aug 07, 2025Abstract:Natural language explanations in recommender systems are often framed as a review generation task, leveraging user reviews as ground-truth supervision. While convenient, this approach conflates a user's opinion with the system's reasoning, leading to explanations that may be fluent but fail to reflect the true logic behind recommendations. In this work, we revisit the core objective of explainable recommendation: to transparently communicate why an item is recommended by linking user needs to relevant item features. Through a comprehensive analysis of existing methods across multiple benchmark datasets, we identify common limitations-explanations that are weakly aligned with model predictions, vague or inaccurate in identifying user intents, and overly repetitive or generic. To overcome these challenges, we propose FIRE, a lightweight and interpretable framework that combines SHAP-based feature attribution with structured, prompt-driven language generation. FIRE produces faithful, diverse, and user-aligned explanations, grounded in the actual decision-making process of the model. Our results demonstrate that FIRE not only achieves competitive recommendation accuracy but also significantly improves explanation quality along critical dimensions such as alignment, structure, and faithfulness. This work highlights the need to move beyond the review-as-explanation paradigm and toward explanation methods that are both accountable and interpretable.
Log-Sum-Exponential Estimator for Off-Policy Evaluation and Learning
Jun 07, 2025Abstract:Off-policy learning and evaluation leverage logged bandit feedback datasets, which contain context, action, propensity score, and feedback for each data point. These scenarios face significant challenges due to high variance and poor performance with low-quality propensity scores and heavy-tailed reward distributions. We address these issues by introducing a novel estimator based on the log-sum-exponential (LSE) operator, which outperforms traditional inverse propensity score estimators. Our LSE estimator demonstrates variance reduction and robustness under heavy-tailed conditions. For off-policy evaluation, we derive upper bounds on the estimator's bias and variance. In the off-policy learning scenario, we establish bounds on the regret -- the performance gap between our LSE estimator and the optimal policy -- assuming bounded $(1+\epsilon)$-th moment of weighted reward. Notably, we achieve a convergence rate of $O(n^{-\epsilon/(1+ \epsilon)})$ for the regret bounds, where $\epsilon \in [0,1]$ and $n$ is the size of logged bandit feedback dataset. Theoretical analysis is complemented by comprehensive empirical evaluations in both off-policy learning and evaluation scenarios, confirming the practical advantages of our approach. The code for our estimator is available at the following link: https://github.com/armin-behnamnia/lse-offpolicy-learning.
Fast, Not Fancy: Rethinking G2P with Rich Data and Rule-Based Models
May 19, 2025Abstract:Homograph disambiguation remains a significant challenge in grapheme-to-phoneme (G2P) conversion, especially for low-resource languages. This challenge is twofold: (1) creating balanced and comprehensive homograph datasets is labor-intensive and costly, and (2) specific disambiguation strategies introduce additional latency, making them unsuitable for real-time applications such as screen readers and other accessibility tools. In this paper, we address both issues. First, we propose a semi-automated pipeline for constructing homograph-focused datasets, introduce the HomoRich dataset generated through this pipeline, and demonstrate its effectiveness by applying it to enhance a state-of-the-art deep learning-based G2P system for Persian. Second, we advocate for a paradigm shift - utilizing rich offline datasets to inform the development of fast, rule-based methods suitable for latency-sensitive accessibility applications like screen readers. To this end, we improve one of the most well-known rule-based G2P systems, eSpeak, into a fast homograph-aware version, HomoFast eSpeak. Our results show an approximate 30% improvement in homograph disambiguation accuracy for the deep learning-based and eSpeak systems.
Generative AI for Character Animation: A Comprehensive Survey of Techniques, Applications, and Future Directions
Apr 27, 2025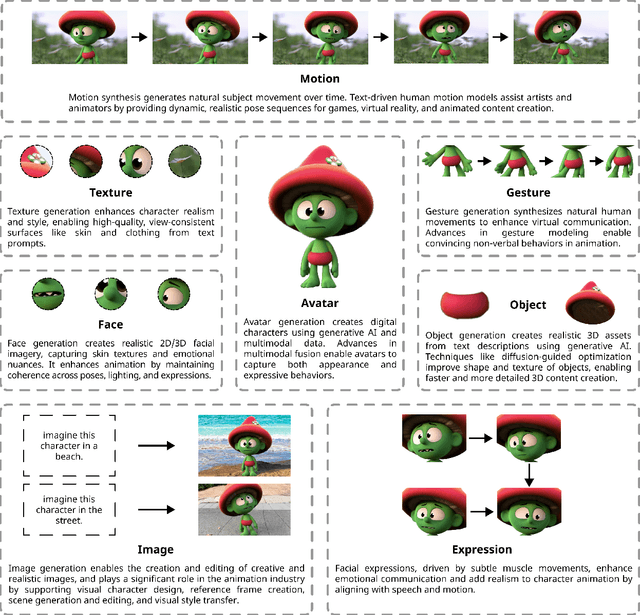
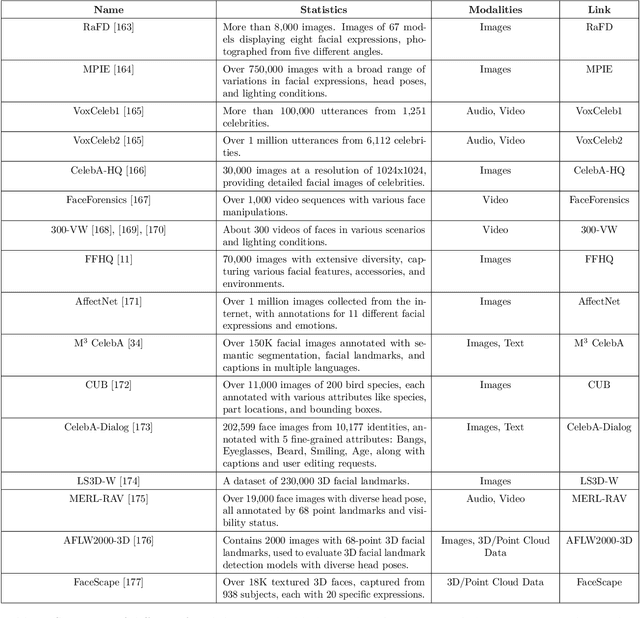
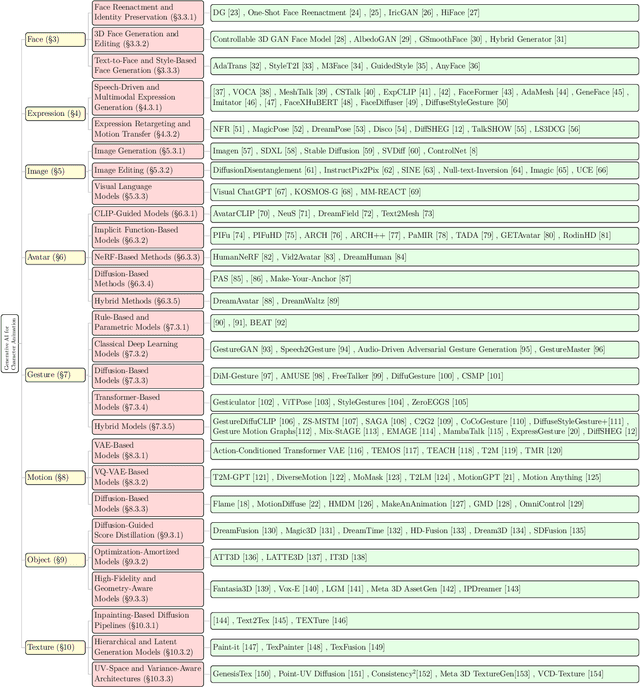
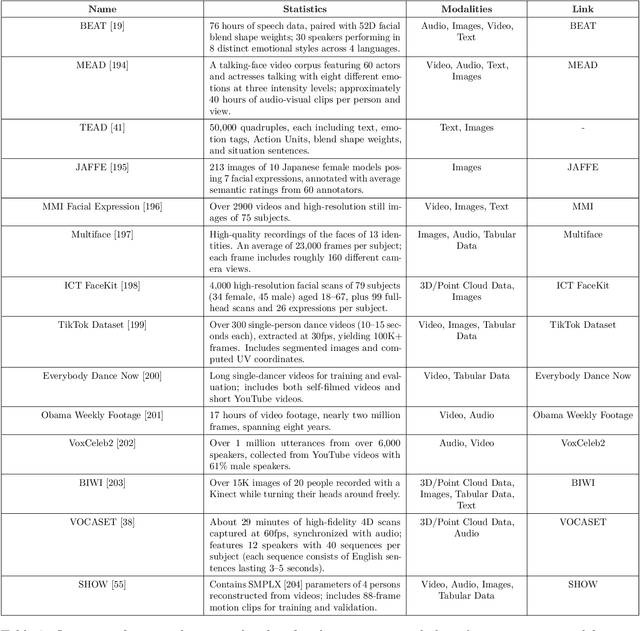
Abstract:Generative AI is reshaping art, gaming, and most notably animation. Recent breakthroughs in foundation and diffusion models have reduced the time and cost of producing animated content. Characters are central animation components, involving motion, emotions, gestures, and facial expressions. The pace and breadth of advances in recent months make it difficult to maintain a coherent view of the field, motivating the need for an integrative review. Unlike earlier overviews that treat avatars, gestures, or facial animation in isolation, this survey offers a single, comprehensive perspective on all the main generative AI applications for character animation. We begin by examining the state-of-the-art in facial animation, expression rendering, image synthesis, avatar creation, gesture modeling, motion synthesis, object generation, and texture synthesis. We highlight leading research, practical deployments, commonly used datasets, and emerging trends for each area. To support newcomers, we also provide a comprehensive background section that introduces foundational models and evaluation metrics, equipping readers with the knowledge needed to enter the field. We discuss open challenges and map future research directions, providing a roadmap to advance AI-driven character-animation technologies. This survey is intended as a resource for researchers and developers entering the field of generative AI animation or adjacent fields. Resources are available at: https://github.com/llm-lab-org/Generative-AI-for-Character-Animation-Survey.
Epistemic Uncertainty-aware Recommendation Systems via Bayesian Deep Ensemble Learning
Apr 14, 2025



Abstract:Recommending items to users has long been a fundamental task, and studies have tried to improve it ever since. Most well-known models commonly employ representation learning to map users and items into a unified embedding space for matching assessment. These approaches have primary limitations, especially when dealing with explicit feedback and sparse data contexts. Two primary limitations are their proneness to overfitting and failure to incorporate epistemic uncertainty in predictions. To address these problems, we propose a novel Bayesian Deep Ensemble Collaborative Filtering method named BDECF. To improve model generalization and quality, we utilize Bayesian Neural Networks, which incorporate uncertainty within their weight parameters. In addition, we introduce a new interpretable non-linear matching approach for the user and item embeddings, leveraging the advantages of the attention mechanism. Furthermore, we endorse the implementation of an ensemble-based supermodel to generate more robust and reliable predictions, resulting in a more complete model. Empirical evaluation through extensive experiments and ablation studies across a range of publicly accessible real-world datasets with differing sparsity characteristics confirms our proposed method's effectiveness and the importance of its components.
Momentum-based Distributed Resource Scheduling Optimization Subject to Sector-Bound Nonlinearity and Latency
Mar 08, 2025Abstract:This paper proposes an accelerated consensus-based distributed iterative algorithm for resource allocation and scheduling. The proposed gradient-tracking algorithm introduces an auxiliary variable to add momentum towards the optimal state. We prove that this solution is all-time feasible, implying that the coupling constraint always holds along the algorithm iterative procedure; therefore, the algorithm can be terminated at any time. This is in contrast to the ADMM-based solutions that meet constraint feasibility asymptotically. Further, we show that the proposed algorithm can handle possible link nonlinearity due to logarithmically-quantized data transmission (or any sign-preserving odd sector-bound nonlinear mapping). We prove convergence over uniformly-connected dynamic networks (i.e., a hybrid setup) that may occur in mobile and time-varying multi-agent networks. Further, the latency issue over the network is addressed by proposing delay-tolerant solutions. To our best knowledge, accelerated momentum-based convergence, nonlinear linking, all-time feasibility, uniform network connectivity, and handling (possible) time delays are not altogether addressed in the literature. These contributions make our solution practical in many real-world applications.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge