Arash Afkanpour
Automated Capability Evaluation of Foundation Models
May 22, 2025Abstract:Current evaluation frameworks for foundation models rely heavily on fixed, manually curated benchmarks, limiting their ability to capture the full breadth of model capabilities. This paper introduces Active learning for Capability Evaluation (ACE), a novel framework for scalable, automated, and fine-grained evaluation of foundation models. ACE leverages the knowledge embedded in powerful language models to decompose a domain into semantically meaningful capabilities and generate diverse evaluation tasks, significantly reducing human effort. To maximize coverage and efficiency, ACE models a subject model's performance as a capability function over a latent semantic space and uses active learning to prioritize the evaluation of the most informative capabilities. This adaptive evaluation strategy enables cost-effective discovery of strengths, weaknesses, and failure modes that static benchmarks may miss. Our results suggest that ACE provides a more complete and informative picture of model capabilities, which is essential for safe and well-informed deployment of foundation models.
Advancing Medical Representation Learning Through High-Quality Data
Mar 18, 2025Abstract:Despite the growing scale of medical Vision-Language datasets, the impact of dataset quality on model performance remains under-explored. We introduce Open-PMC, a high-quality medical dataset from PubMed Central, containing 2.2 million image-text pairs, enriched with image modality annotations, subfigures, and summarized in-text references. Notably, the in-text references provide richer medical context, extending beyond the abstract information typically found in captions. Through extensive experiments, we benchmark Open-PMC against larger datasets across retrieval and zero-shot classification tasks. Our results show that dataset quality-not just size-drives significant performance gains. We complement our benchmark with an in-depth analysis of feature representation. Our findings highlight the crucial role of data curation quality in advancing multimodal medical AI. We release Open-PMC, along with the trained models and our codebase.
A Shared Encoder Approach to Multimodal Representation Learning
Mar 03, 2025Abstract:Multimodal representation learning has demonstrated remarkable potential in enabling models to process and integrate diverse data modalities, such as text and images, for improved understanding and performance. While the medical domain can benefit significantly from this paradigm, the scarcity of paired multimodal data and reliance on proprietary or pretrained encoders pose significant challenges. In this work, we present a shared encoder framework for multimodal representation learning tailored to the medical domain. Our approach employs a single set of encoder parameters shared across modalities, augmented with learnable modality features. Empirical results demonstrate that our shared encoder idea achieves superior performance compared to separate modality-specific encoders, demonstrating improved generalization in data-constrained settings. Notably, the performance gains are more pronounced with fewer training examples, underscoring the efficiency of our shared encoder framework for real-world medical applications with limited data. Our code and experiment setup are available at https://github.com/VectorInstitute/shared_encoder.
Benchmarking Vision-Language Contrastive Methods for Medical Representation Learning
Jun 11, 2024



Abstract:We perform a comprehensive benchmarking of contrastive frameworks for learning multimodal representations in the medical domain. Through this study, we aim to answer the following research questions: (i) How transferable are general-domain representations to the medical domain? (ii) Is multimodal contrastive training sufficient, or does it benefit from unimodal training as well? (iii) What is the impact of feature granularity on the effectiveness of multimodal medical representation learning? To answer these questions, we investigate eight contrastive learning approaches under identical training setups, and train them on 2.8 million image-text pairs from four datasets, and evaluate them on 25 downstream tasks, including classification (zero-shot and linear probing), image-to-text and text-to-image retrieval, and visual question-answering. Our findings suggest a positive answer to the first question, a negative answer to the second question, and the benefit of learning fine-grained features. Finally, we make our code publicly available.
Few-shot Tuning of Foundation Models for Class-incremental Learning
May 26, 2024Abstract:For the first time, we explore few-shot tuning of vision foundation models for class-incremental learning. Unlike existing few-shot class incremental learning (FSCIL) methods, which train an encoder on a base session to ensure forward compatibility for future continual learning, foundation models are generally trained on large unlabelled data without such considerations. This renders prior methods from traditional FSCIL incompatible for FSCIL with the foundation model. To this end, we propose Consistency-guided Asynchronous Contrastive Tuning (CoACT), a new approach to continually tune foundation models for new classes in few-shot settings. CoACT comprises three components: (i) asynchronous contrastive tuning, which learns new classes by including LoRA modules in the pre-trained encoder, while enforcing consistency between two asynchronous encoders; (ii) controlled fine-tuning, which facilitates effective tuning of a subset of the foundation model; and (iii) consistency-guided incremental tuning, which enforces additional regularization during later sessions to reduce forgetting of the learned classes. We perform an extensive study on 16 diverse datasets and demonstrate the effectiveness of CoACT, outperforming the best baseline method by 2.47% on average and with up to 12.52% on individual datasets. Additionally, CoACT shows reduced forgetting and robustness in low-shot experiments. As an added bonus, CoACT shows up to 13.5% improvement in standard FSCIL over the current SOTA on benchmark evaluations. We make our code publicly available at https://github.com/ShuvenduRoy/CoACT-FSCIL.
EHRMamba: Towards Generalizable and Scalable Foundation Models for Electronic Health Records
May 23, 2024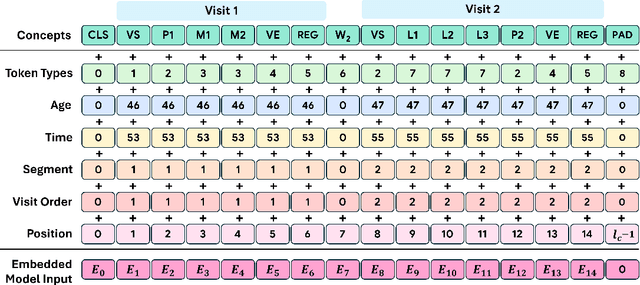
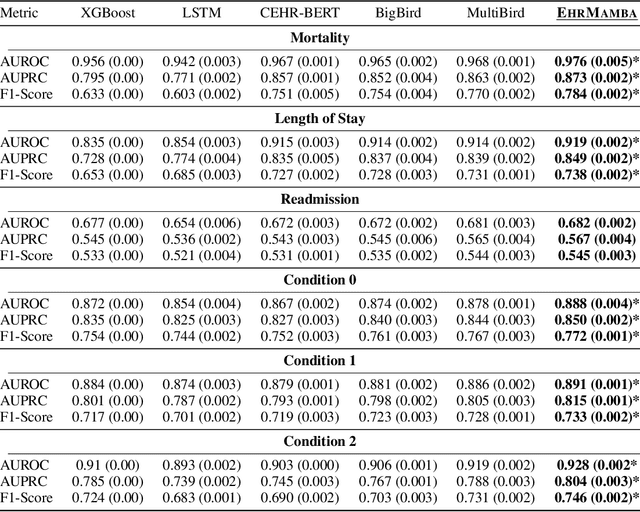
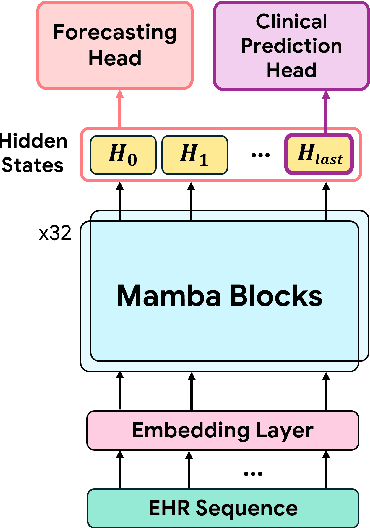
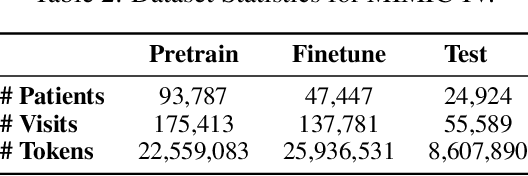
Abstract:Transformers have significantly advanced the modeling of Electronic Health Records (EHR), yet their deployment in real-world healthcare is limited by several key challenges. Firstly, the quadratic computational cost and insufficient context length of these models pose significant obstacles for hospitals in processing the extensive medical histories typical in EHR data. Additionally, existing models employ separate finetuning for each clinical task, complicating maintenance in healthcare environments. Moreover, these models focus exclusively on either clinical prediction or EHR forecasting, lacking the flexibility to perform well across both. To overcome these limitations, we introduce EHRMamba, a robust foundation model built on the Mamba architecture. EHRMamba can process sequences up to four times longer than previous models due to its linear computational cost. We also introduce a novel approach to Multitask Prompted Finetuning (MTF) for EHR data, which enables EHRMamba to simultaneously learn multiple clinical tasks in a single finetuning phase, significantly enhancing deployment and cross-task generalization. Furthermore, our model leverages the HL7 FHIR data standard to simplify integration into existing hospital systems. Alongside EHRMamba, we open-source Odyssey, a toolkit designed to support the development and deployment of EHR foundation models, with an emphasis on data standardization and interpretability. Our evaluations on the MIMIC-IV dataset demonstrate that EHRMamba advances state-of-the-art performance across 6 major clinical tasks and excels in EHR forecasting, marking a significant leap forward in the field.
Can Generative Models Improve Self-Supervised Representation Learning?
Mar 09, 2024



Abstract:The rapid advancement in self-supervised learning (SSL) has highlighted its potential to leverage unlabeled data for learning powerful visual representations. However, existing SSL approaches, particularly those employing different views of the same image, often rely on a limited set of predefined data augmentations. This constrains the diversity and quality of transformations, which leads to sub-optimal representations. In this paper, we introduce a novel framework that enriches the SSL paradigm by utilizing generative models to produce semantically consistent image augmentations. By directly conditioning generative models on a source image representation, our method enables the generation of diverse augmentations while maintaining the semantics of the source image, thus offering a richer set of data for self-supervised learning. Our experimental results demonstrate that our framework significantly enhances the quality of learned visual representations. This research demonstrates that incorporating generative models into the SSL workflow opens new avenues for exploring the potential of unlabeled visual data. This development paves the way for more robust and versatile representation learning techniques.
Random Field Augmentations for Self-Supervised Representation Learning
Nov 07, 2023Abstract:Self-supervised representation learning is heavily dependent on data augmentations to specify the invariances encoded in representations. Previous work has shown that applying diverse data augmentations is crucial to downstream performance, but augmentation techniques remain under-explored. In this work, we propose a new family of local transformations based on Gaussian random fields to generate image augmentations for self-supervised representation learning. These transformations generalize the well-established affine and color transformations (translation, rotation, color jitter, etc.) and greatly increase the space of augmentations by allowing transformation parameter values to vary from pixel to pixel. The parameters are treated as continuous functions of spatial coordinates, and modeled as independent Gaussian random fields. Empirical results show the effectiveness of the new transformations for self-supervised representation learning. Specifically, we achieve a 1.7% top-1 accuracy improvement over baseline on ImageNet downstream classification, and a 3.6% improvement on out-of-distribution iNaturalist downstream classification. However, due to the flexibility of the new transformations, learned representations are sensitive to hyperparameters. While mild transformations improve representations, we observe that strong transformations can degrade the structure of an image, indicating that balancing the diversity and strength of augmentations is important for improving generalization of learned representations.
Federated Variational Inference: Towards Improved Personalization and Generalization
May 25, 2023



Abstract:Conventional federated learning algorithms train a single global model by leveraging all participating clients' data. However, due to heterogeneity in client generative distributions and predictive models, these approaches may not appropriately approximate the predictive process, converge to an optimal state, or generalize to new clients. We study personalization and generalization in stateless cross-device federated learning setups assuming heterogeneity in client data distributions and predictive models. We first propose a hierarchical generative model and formalize it using Bayesian Inference. We then approximate this process using Variational Inference to train our model efficiently. We call this algorithm Federated Variational Inference (FedVI). We use PAC-Bayes analysis to provide generalization bounds for FedVI. We evaluate our model on FEMNIST and CIFAR-100 image classification and show that FedVI beats the state-of-the-art on both tasks.
BERT for Long Documents: A Case Study of Automated ICD Coding
Nov 04, 2022



Abstract:Transformer models have achieved great success across many NLP problems. However, previous studies in automated ICD coding concluded that these models fail to outperform some of the earlier solutions such as CNN-based models. In this paper we challenge this conclusion. We present a simple and scalable method to process long text with the existing transformer models such as BERT. We show that this method significantly improves the previous results reported for transformer models in ICD coding, and is able to outperform one of the prominent CNN-based methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge