Soroush Farghadani
IA-GCN: Interpretable Attention based Graph Convolutional Network for Disease prediction
Mar 29, 2021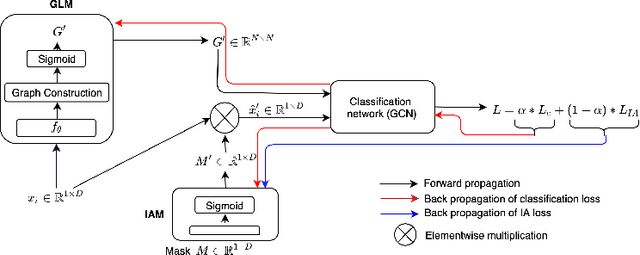
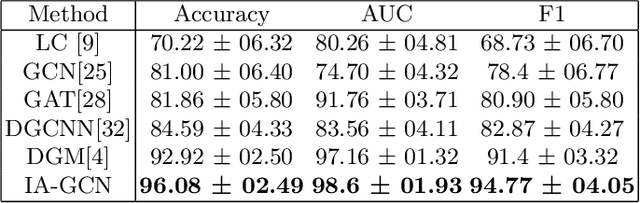
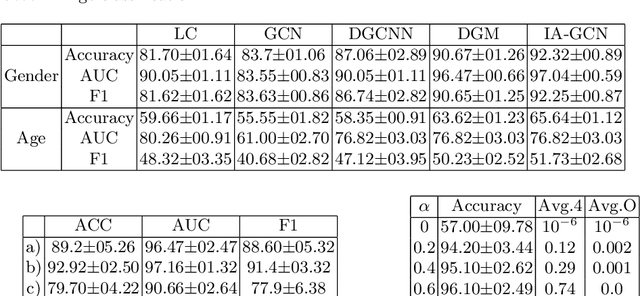
Abstract:Interpretability in Graph Convolutional Networks (GCNs) has been explored to some extent in computer vision in general, yet, in the medical domain, it requires further examination. Moreover, most of the interpretability approaches for GCNs, especially in the medical domain, focus on interpreting the model in a post hoc fashion. In this paper, we propose an interpretable graph learning-based model which 1) interprets the clinical relevance of the input features towards the task, 2) uses the explanation to improve the model performance and, 3) learns a population level latent graph that may be used to interpret the cohort's behavior. In a clinical scenario, such a model can assist the clinical experts in better decision-making for diagnosis and treatment planning. The main novelty lies in the interpretable attention module (IAM), which directly operates on multi-modal features. Our IAM learns the attention for each feature based on the unique interpretability-specific losses. We show the application on two publicly available datasets, Tadpole and UKBB, for three tasks of disease, age, and gender prediction. Our proposed model shows superior performance with respect to compared methods with an increase in an average accuracy of 3.2% for Tadpole, 1.6% for UKBB Gender, and 2% for the UKBB Age prediction task. Further, we show exhaustive validation and clinical interpretation of our results.
Dementia Severity Classification under Small Sample Size and Weak Supervision in Thick Slice MRI
Mar 18, 2021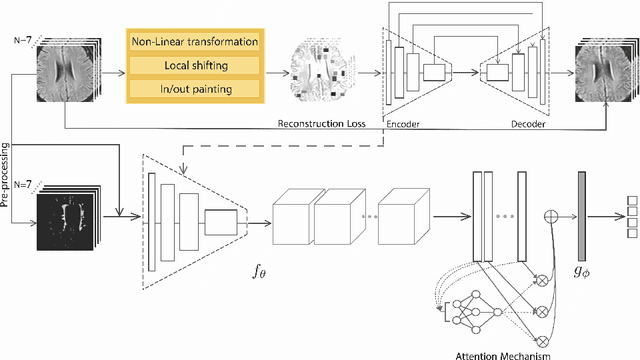
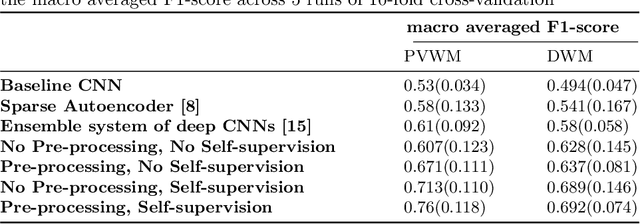
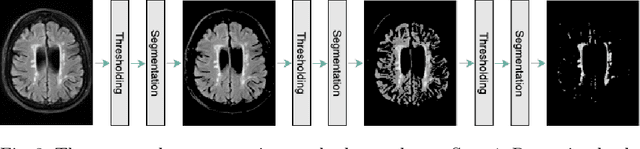
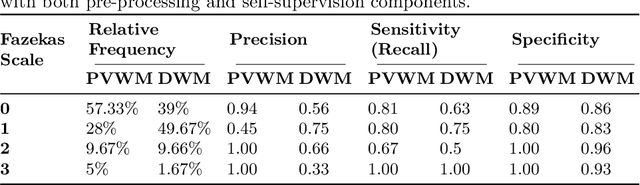
Abstract:Early detection of dementia through specific biomarkers in MR images plays a critical role in developing support strategies proactively. Fazekas scale facilitates an accurate quantitative assessment of the severity of white matter lesions and hence the disease. Imaging Biomarkers of dementia are multiple and comprehensive documentation of them is time-consuming. Therefore, any effort to automatically extract these biomarkers will be of clinical value while reducing inter-rater discrepancies. To tackle this problem, we propose to classify the disease severity based on the Fazekas scale through the visual biomarkers, namely the Periventricular White Matter (PVWM) and the Deep White Matter (DWM) changes, in the real-world setting of thick-slice MRI. Small training sample size and weak supervision in form of assigning severity labels to the whole MRI stack are among the main challenges. To combat the mentioned issues, we have developed a deep learning pipeline that employs self-supervised representation learning, multiple instance learning, and appropriate pre-processing steps. We use pretext tasks such as non-linear transformation, local shuffling, in- and out-painting for self-supervised learning of useful features in this domain. Furthermore, an attention model is used to determine the relevance of each MRI slice for predicting the Fazekas scale in an unsupervised manner. We show the significant superiority of our method in distinguishing different classes of dementia compared to state-of-the-art methods in our mentioned setting, which improves the macro averaged F1-score of state-of-the-art from 61% to 76% in PVWM, and from 58% to 69.2% in DWM.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge