Anees Kazi
Computer Aided Medical Procedures, Technische Universit Munchen, Germany
The Importance of Model Inspection for Better Understanding Performance Characteristics of Graph Neural Networks
May 02, 2024


Abstract:This study highlights the importance of conducting comprehensive model inspection as part of comparative performance analyses. Here, we investigate the effect of modelling choices on the feature learning characteristics of graph neural networks applied to a brain shape classification task. Specifically, we analyse the effect of using parameter-efficient, shared graph convolutional submodels compared to structure-specific, non-shared submodels. Further, we assess the effect of mesh registration as part of the data harmonisation pipeline. We find substantial differences in the feature embeddings at different layers of the models. Our results highlight that test accuracy alone is insufficient to identify important model characteristics such as encoded biases related to data source or potentially non-discriminative features learned in submodels. Our model inspection framework offers a valuable tool for practitioners to better understand performance characteristics of deep learning models in medical imaging.
On Discprecncies between Perturbation Evaluations of Graph Neural Network Attributions
Jan 01, 2024Abstract:Neural networks are increasingly finding their way into the realm of graphs and modeling relationships between features. Concurrently graph neural network explanation approaches are being invented to uncover relationships between the nodes of the graphs. However, there is a disparity between the existing attribution methods, and it is unclear which attribution to trust. Therefore research has introduced evaluation experiments that assess them from different perspectives. In this work, we assess attribution methods from a perspective not previously explored in the graph domain: retraining. The core idea is to retrain the network on important (or not important) relationships as identified by the attributions and evaluate how networks can generalize based on these relationships. We reformulate the retraining framework to sidestep issues lurking in the previous formulation and propose guidelines for correct analysis. We run our analysis on four state-of-the-art GNN attribution methods and five synthetic and real-world graph classification datasets. The analysis reveals that attributions perform variably depending on the dataset and the network. Most importantly, we observe that the famous GNNExplainer performs similarly to an arbitrary designation of edge importance. The study concludes that the retraining evaluation cannot be used as a generalized benchmark and recommends it as a toolset to evaluate attributions on a specifically addressed network, dataset, and sparsity.
Multi-Head Graph Convolutional Network for Structural Connectome Classification
May 02, 2023Abstract:We tackle classification based on brain connectivity derived from diffusion magnetic resonance images. We propose a machine-learning model inspired by graph convolutional networks (GCNs), which takes a brain connectivity input graph and processes the data separately through a parallel GCN mechanism with multiple heads. The proposed network is a simple design that employs different heads involving graph convolutions focused on edges and nodes, capturing representations from the input data thoroughly. To test the ability of our model to extract complementary and representative features from brain connectivity data, we chose the task of sex classification. This quantifies the degree to which the connectome varies depending on the sex, which is important for improving our understanding of health and disease in both sexes. We show experiments on two publicly available datasets: PREVENT-AD (347 subjects) and OASIS3 (771 subjects). The proposed model demonstrates the highest performance compared to the existing machine-learning algorithms we tested, including classical methods and (graph and non-graph) deep learning. We provide a detailed analysis of each component of our model.
Latent Graph Inference using Product Manifolds
Nov 26, 2022Abstract:Graph Neural Networks usually rely on the assumption that the graph topology is available to the network as well as optimal for the downstream task. Latent graph inference allows models to dynamically learn the intrinsic graph structure of problems where the connectivity patterns of data may not be directly accessible. In this work, we generalize the discrete Differentiable Graph Module (dDGM) for latent graph learning. The original dDGM architecture used the Euclidean plane to encode latent features based on which the latent graphs were generated. By incorporating Riemannian geometry into the model and generating more complex embedding spaces, we can improve the performance of the latent graph inference system. In particular, we propose a computationally tractable approach to produce product manifolds of constant curvature model spaces that can encode latent features of varying structure. The latent representations mapped onto the inferred product manifold are used to compute richer similarity measures that are leveraged by the latent graph learning model to obtain optimized latent graphs. Moreover, the curvature of the product manifold is learned during training alongside the rest of the network parameters and based on the downstream task, rather than it being a static embedding space. Our novel approach is tested on a wide range of datasets, and outperforms the original dDGM model.
Unsupervised pre-training of graph transformers on patient population graphs
Jul 21, 2022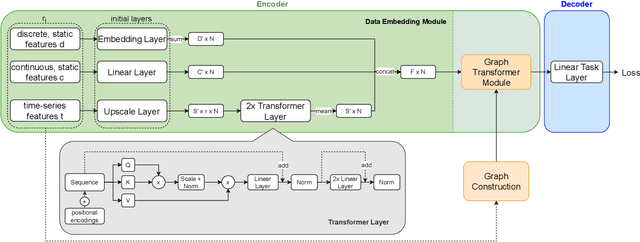
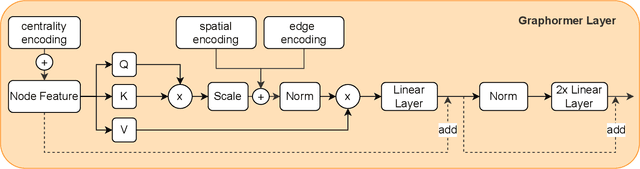
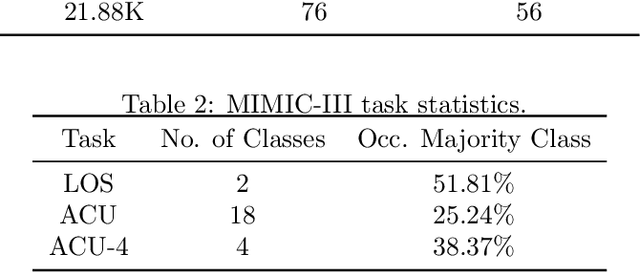
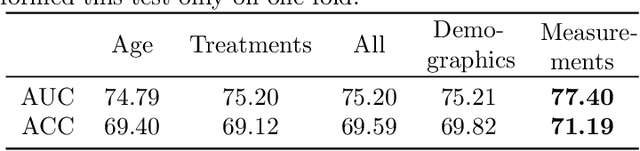
Abstract:Pre-training has shown success in different areas of machine learning, such as Computer Vision, Natural Language Processing (NLP), and medical imaging. However, it has not been fully explored for clinical data analysis. An immense amount of clinical records are recorded, but still, data and labels can be scarce for data collected in small hospitals or dealing with rare diseases. In such scenarios, pre-training on a larger set of unlabelled clinical data could improve performance. In this paper, we propose novel unsupervised pre-training techniques designed for heterogeneous, multi-modal clinical data for patient outcome prediction inspired by masked language modeling (MLM), by leveraging graph deep learning over population graphs. To this end, we further propose a graph-transformer-based network, designed to handle heterogeneous clinical data. By combining masking-based pre-training with a transformer-based network, we translate the success of masking-based pre-training in other domains to heterogeneous clinical data. We show the benefit of our pre-training method in a self-supervised and a transfer learning setting, utilizing three medical datasets TADPOLE, MIMIC-III, and a Sepsis Prediction Dataset. We find that our proposed pre-training methods help in modeling the data at a patient and population level and improve performance in different fine-tuning tasks on all datasets.
Graph-in-Graph (GiG): Learning interpretable latent graphs in non-Euclidean domain for biological and healthcare applications
Apr 01, 2022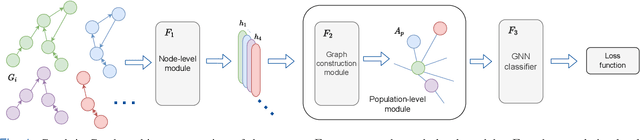
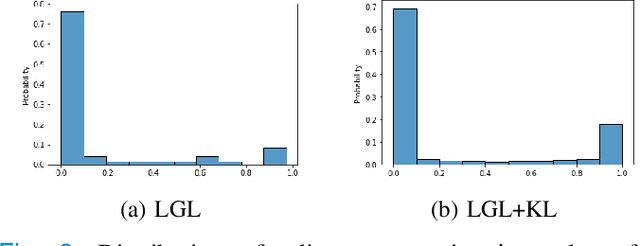
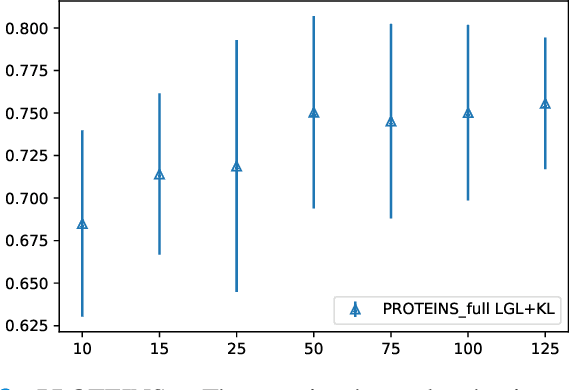
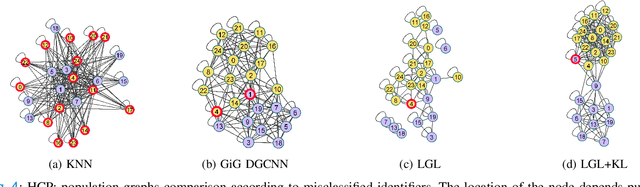
Abstract:Graphs are a powerful tool for representing and analyzing unstructured, non-Euclidean data ubiquitous in the healthcare domain. Two prominent examples are molecule property prediction and brain connectome analysis. Importantly, recent works have shown that considering relationships between input data samples have a positive regularizing effect for the downstream task in healthcare applications. These relationships are naturally modeled by a (possibly unknown) graph structure between input samples. In this work, we propose Graph-in-Graph (GiG), a neural network architecture for protein classification and brain imaging applications that exploits the graph representation of the input data samples and their latent relation. We assume an initially unknown latent-graph structure between graph-valued input data and propose to learn end-to-end a parametric model for message passing within and across input graph samples, along with the latent structure connecting the input graphs. Further, we introduce a degree distribution loss that helps regularize the predicted latent relationships structure. This regularization can significantly improve the downstream task. Moreover, the obtained latent graph can represent patient population models or networks of molecule clusters, providing a level of interpretability and knowledge discovery in the input domain of particular value in healthcare.
Unsupervised Pre-Training on Patient Population Graphs for Patient-Level Predictions
Mar 23, 2022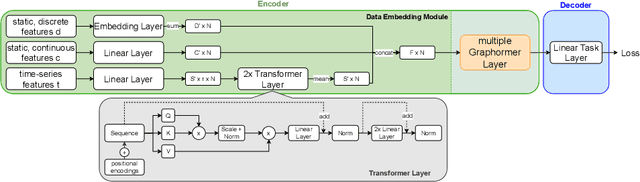
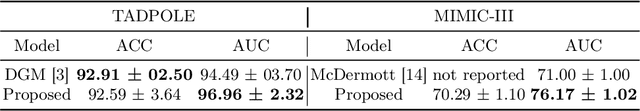
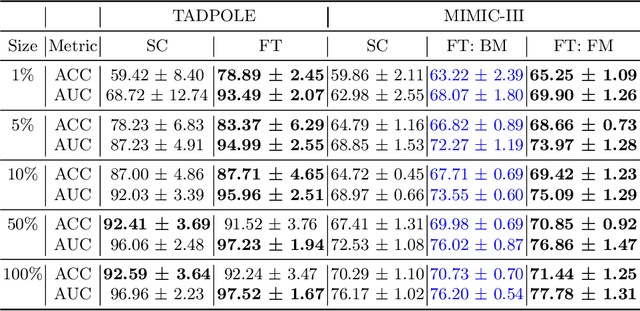
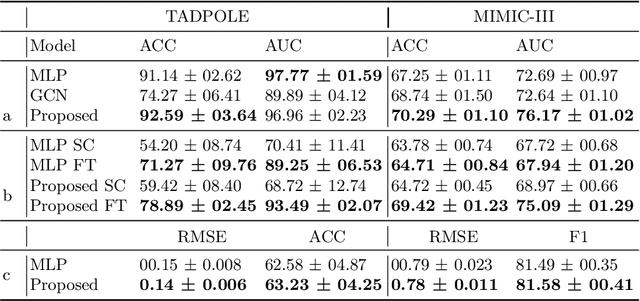
Abstract:Pre-training has shown success in different areas of machine learning, such as Computer Vision (CV), Natural Language Processing (NLP) and medical imaging. However, it has not been fully explored for clinical data analysis. Even though an immense amount of Electronic Health Record (EHR) data is recorded, data and labels can be scarce if the data is collected in small hospitals or deals with rare diseases. In such scenarios, pre-training on a larger set of EHR data could improve the model performance. In this paper, we apply unsupervised pre-training to heterogeneous, multi-modal EHR data for patient outcome prediction. To model this data, we leverage graph deep learning over population graphs. We first design a network architecture based on graph transformer designed to handle various input feature types occurring in EHR data, like continuous, discrete, and time-series features, allowing better multi-modal data fusion. Further, we design pre-training methods based on masked imputation to pre-train our network before fine-tuning on different end tasks. Pre-training is done in a fully unsupervised fashion, which lays the groundwork for pre-training on large public datasets with different tasks and similar modalities in the future. We test our method on two medical datasets of patient records, TADPOLE and MIMIC-III, including imaging and non-imaging features and different prediction tasks. We find that our proposed graph based pre-training method helps in modeling the data at a population level and further improves performance on the fine tuning tasks in terms of AUC on average by 4.15% for MIMIC and 7.64% for TADPOLE.
GKD: Semi-supervised Graph Knowledge Distillation for Graph-Independent Inference
Apr 08, 2021



Abstract:The increased amount of multi-modal medical data has opened the opportunities to simultaneously process various modalities such as imaging and non-imaging data to gain a comprehensive insight into the disease prediction domain. Recent studies using Graph Convolutional Networks (GCNs) provide novel semi-supervised approaches for integrating heterogeneous modalities while investigating the patients' associations for disease prediction. However, when the meta-data used for graph construction is not available at inference time (e.g., coming from a distinct population), the conventional methods exhibit poor performance. To address this issue, we propose a novel semi-supervised approach named GKD based on knowledge distillation. We train a teacher component that employs the label-propagation algorithm besides a deep neural network to benefit from the graph and non-graph modalities only in the training phase. The teacher component embeds all the available information into the soft pseudo-labels. The soft pseudo-labels are then used to train a deep student network for disease prediction of unseen test data for which the graph modality is unavailable. We perform our experiments on two public datasets for diagnosing Autism spectrum disorder, and Alzheimer's disease, along with a thorough analysis on synthetic multi-modal datasets. According to these experiments, GKD outperforms the previous graph-based deep learning methods in terms of accuracy, AUC, and Macro F1.
IA-GCN: Interpretable Attention based Graph Convolutional Network for Disease prediction
Mar 29, 2021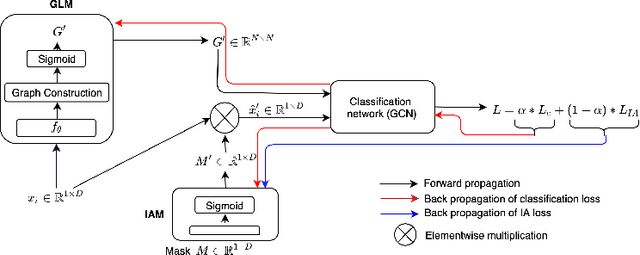
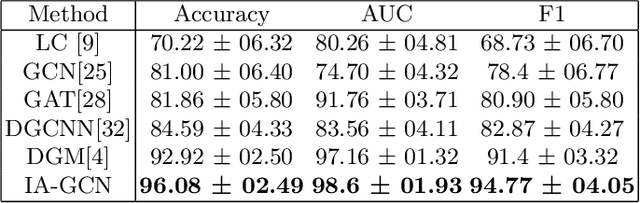
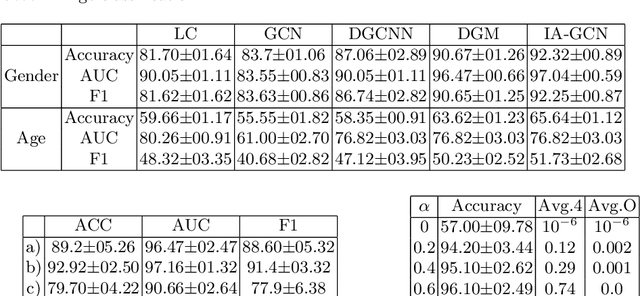
Abstract:Interpretability in Graph Convolutional Networks (GCNs) has been explored to some extent in computer vision in general, yet, in the medical domain, it requires further examination. Moreover, most of the interpretability approaches for GCNs, especially in the medical domain, focus on interpreting the model in a post hoc fashion. In this paper, we propose an interpretable graph learning-based model which 1) interprets the clinical relevance of the input features towards the task, 2) uses the explanation to improve the model performance and, 3) learns a population level latent graph that may be used to interpret the cohort's behavior. In a clinical scenario, such a model can assist the clinical experts in better decision-making for diagnosis and treatment planning. The main novelty lies in the interpretable attention module (IAM), which directly operates on multi-modal features. Our IAM learns the attention for each feature based on the unique interpretability-specific losses. We show the application on two publicly available datasets, Tadpole and UKBB, for three tasks of disease, age, and gender prediction. Our proposed model shows superior performance with respect to compared methods with an increase in an average accuracy of 3.2% for Tadpole, 1.6% for UKBB Gender, and 2% for the UKBB Age prediction task. Further, we show exhaustive validation and clinical interpretation of our results.
RA-GCN: Graph Convolutional Network for Disease Prediction Problems with Imbalanced Data
Feb 27, 2021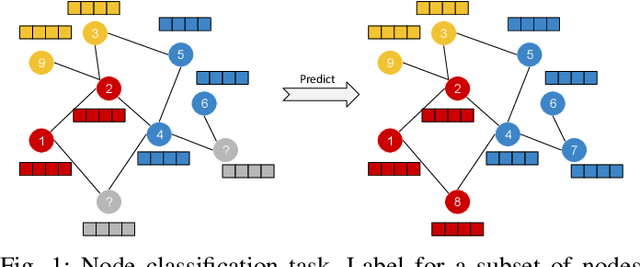
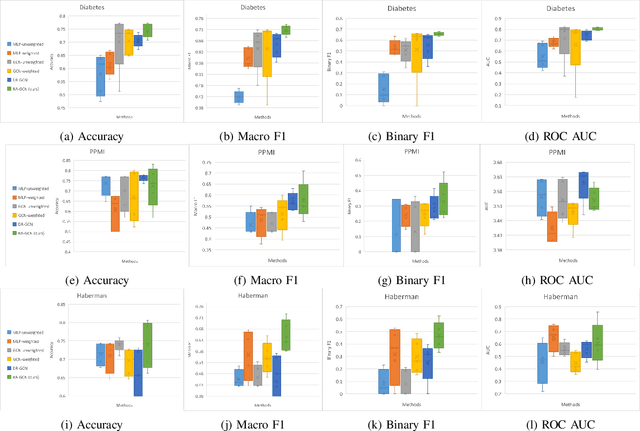
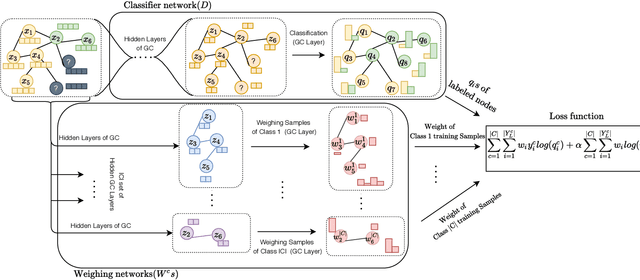
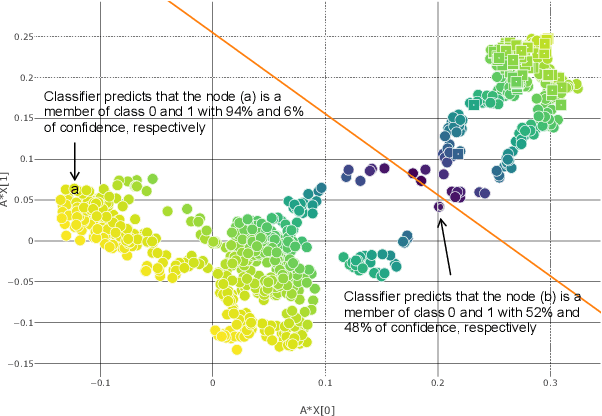
Abstract:Disease prediction is a well-known classification problem in medical applications. Graph neural networks provide a powerful tool for analyzing the patients' features relative to each other. Recently, Graph Convolutional Networks (GCNs) have particularly been studied in the field of disease prediction. Due to the nature of such medical datasets, the class imbalance is a familiar issue in the field of disease prediction. When the class imbalance is present in the data, the existing graph-based classifiers tend to be biased towards the major class(es). Meanwhile, the correct diagnosis of the rare true-positive cases among all the patients is vital. In conventional methods, such imbalance is tackled by assigning appropriate weights to classes in the loss function; however, this solution is still dependent on the relative values of weights, sensitive to outliers, and in some cases biased towards the minor class(es). In this paper, we propose Re-weighted Adversarial Graph Convolutional Network (RA-GCN) to enhance the performance of the graph-based classifier and prevent it from emphasizing the samples of any particular class. This is accomplished by automatically learning to weigh the samples of the classes. For this purpose, a graph-based network is associated with each class, which is responsible for weighing the class samples and informing the classifier about the importance of each sample. Therefore, the classifier adjusts itself and determines the boundary between classes with more attention to the important samples. The parameters of the classifier and weighing networks are trained by an adversarial approach. At the end of the adversarial training process, the boundary of the classifier is more accurate and unbiased. We show the superiority of RA-GCN on synthetic and three publicly available medical datasets compared to the recent method.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge