Kamilia Mullakaeva
Graph-in-Graph (GiG): Learning interpretable latent graphs in non-Euclidean domain for biological and healthcare applications
Apr 01, 2022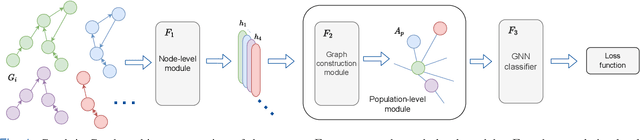
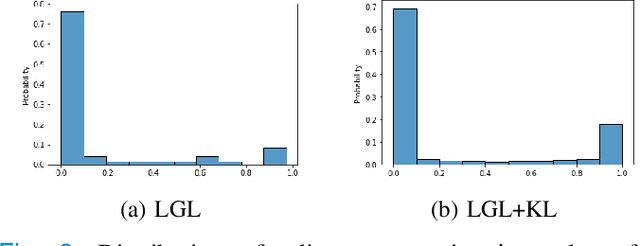
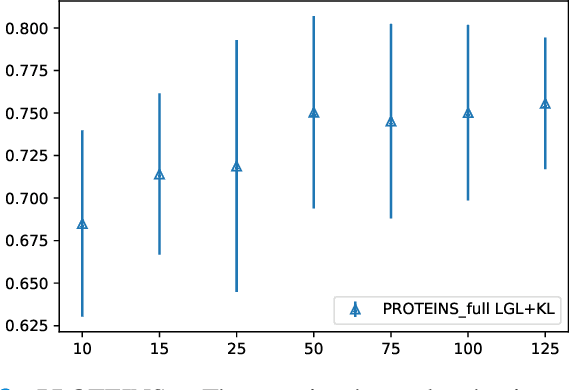
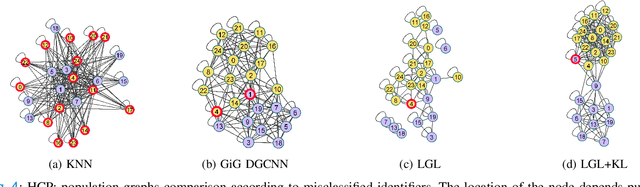
Abstract:Graphs are a powerful tool for representing and analyzing unstructured, non-Euclidean data ubiquitous in the healthcare domain. Two prominent examples are molecule property prediction and brain connectome analysis. Importantly, recent works have shown that considering relationships between input data samples have a positive regularizing effect for the downstream task in healthcare applications. These relationships are naturally modeled by a (possibly unknown) graph structure between input samples. In this work, we propose Graph-in-Graph (GiG), a neural network architecture for protein classification and brain imaging applications that exploits the graph representation of the input data samples and their latent relation. We assume an initially unknown latent-graph structure between graph-valued input data and propose to learn end-to-end a parametric model for message passing within and across input graph samples, along with the latent structure connecting the input graphs. Further, we introduce a degree distribution loss that helps regularize the predicted latent relationships structure. This regularization can significantly improve the downstream task. Moreover, the obtained latent graph can represent patient population models or networks of molecule clusters, providing a level of interpretability and knowledge discovery in the input domain of particular value in healthcare.
Few-shot Structured Radiology Report Generation Using Natural Language Prompts
Mar 29, 2022



Abstract:Chest radiograph reporting is time-consuming, and numerous solutions to automate this process have been proposed. Due to the complexity of medical information, the variety of writing styles, and free text being prone to typos and inconsistencies, the efficacy of quantifying the clinical accuracy of free-text reports using natural language processing measures is challenging. On the other hand, structured reports ensure consistency and can more easily be used as a quality assurance tool. To accomplish this, we present a strategy for predicting clinical observations and their anatomical location that is easily extensible to other structured findings. First, we train a contrastive language-image model using related chest radiographs and free-text radiological reports. Then, we create textual prompts for each structured finding and optimize a classifier for predicting clinical findings and their associations within the medical image. The results indicate that even when only a few image-level annotations are used for training, the method can localize pathologies in chest radiographs and generate structured reports.
Peri-Diagnostic Decision Support Through Cost-Efficient Feature Acquisition at Test-Time
Mar 31, 2020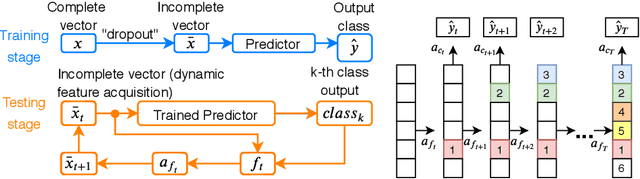
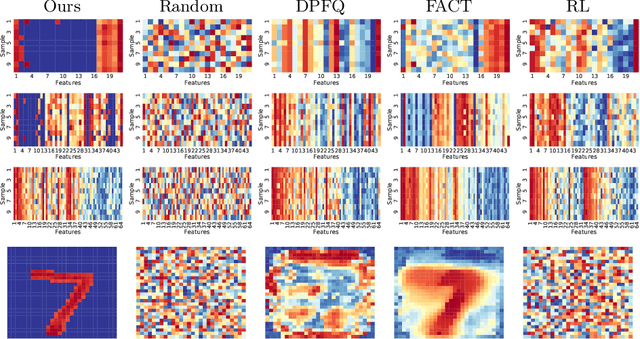
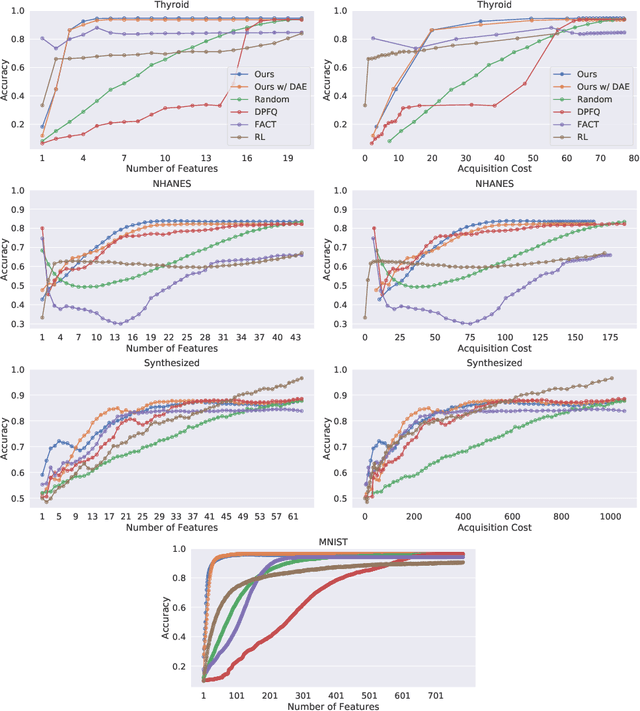
Abstract:Computer-aided diagnosis (CADx) algorithms in medicine provide patient-specific decision support for physicians. These algorithms are usually applied after full acquisition of high-dimensional multimodal examination data, and often assume feature-completeness. This, however, is rarely the case due to examination costs, invasiveness, or a lack of indication. A sub-problem in CADx, which to our knowledge has received very little attention among the CADx community so far, is to guide the physician during the entire peri-diagnostic workflow, including the acquisition stage. We model the following question, asked from a physician's perspective: ''Given the evidence collected so far, which examination should I perform next, in order to achieve the most accurate and efficient diagnostic prediction?''. In this work, we propose a novel approach which is enticingly simple: use dropout at the input layer, and integrated gradients of the trained network at test-time to attribute feature importance dynamically. We validate and explain the effectiveness of our proposed approach using two public medical and two synthetic datasets. Results show that our proposed approach is more cost- and feature-efficient than prior approaches and achieves a higher overall accuracy. This directly translates to less unnecessary examinations for patients, and a quicker, less costly and more accurate decision support for the physician.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge