Mahsa Ghorbani
Intelligent Masking: Deep Q-Learning for Context Encoding in Medical Image Analysis
Apr 04, 2022



Abstract:The need for a large amount of labeled data in the supervised setting has led recent studies to utilize self-supervised learning to pre-train deep neural networks using unlabeled data. Many self-supervised training strategies have been investigated especially for medical datasets to leverage the information available in the much fewer unlabeled data. One of the fundamental strategies in image-based self-supervision is context prediction. In this approach, a model is trained to reconstruct the contents of an arbitrary missing region of an image based on its surroundings. However, the existing methods adopt a random and blind masking approach by focusing uniformly on all regions of the images. This approach results in a lot of unnecessary network updates that cause the model to forget the rich extracted features. In this work, we develop a novel self-supervised approach that occludes targeted regions to improve the pre-training procedure. To this end, we propose a reinforcement learning-based agent which learns to intelligently mask input images through deep Q-learning. We show that training the agent against the prediction model can significantly improve the semantic features extracted for downstream classification tasks. We perform our experiments on two public datasets for diagnosing breast cancer in the ultrasound images and detecting lower-grade glioma with MR images. In our experiments, we show that our novel masking strategy advances the learned features according to the performance on the classification task in terms of accuracy, macro F1, and AUROC.
GKD: Semi-supervised Graph Knowledge Distillation for Graph-Independent Inference
Apr 08, 2021



Abstract:The increased amount of multi-modal medical data has opened the opportunities to simultaneously process various modalities such as imaging and non-imaging data to gain a comprehensive insight into the disease prediction domain. Recent studies using Graph Convolutional Networks (GCNs) provide novel semi-supervised approaches for integrating heterogeneous modalities while investigating the patients' associations for disease prediction. However, when the meta-data used for graph construction is not available at inference time (e.g., coming from a distinct population), the conventional methods exhibit poor performance. To address this issue, we propose a novel semi-supervised approach named GKD based on knowledge distillation. We train a teacher component that employs the label-propagation algorithm besides a deep neural network to benefit from the graph and non-graph modalities only in the training phase. The teacher component embeds all the available information into the soft pseudo-labels. The soft pseudo-labels are then used to train a deep student network for disease prediction of unseen test data for which the graph modality is unavailable. We perform our experiments on two public datasets for diagnosing Autism spectrum disorder, and Alzheimer's disease, along with a thorough analysis on synthetic multi-modal datasets. According to these experiments, GKD outperforms the previous graph-based deep learning methods in terms of accuracy, AUC, and Macro F1.
RA-GCN: Graph Convolutional Network for Disease Prediction Problems with Imbalanced Data
Feb 27, 2021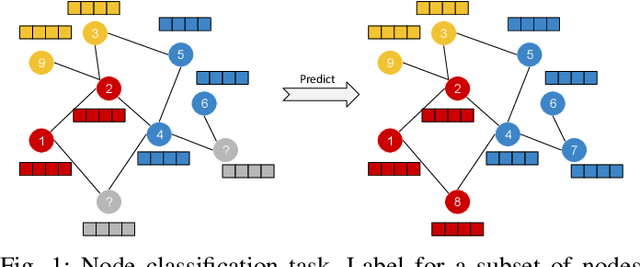
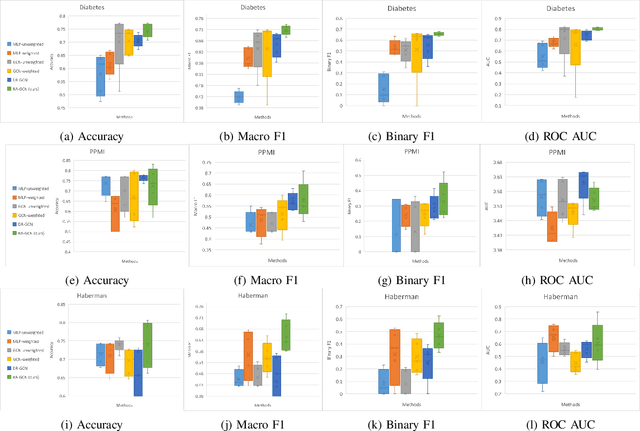
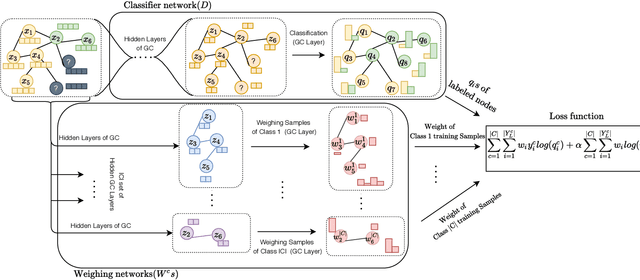
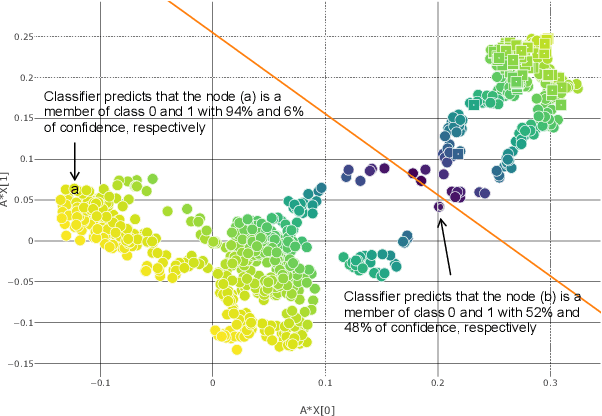
Abstract:Disease prediction is a well-known classification problem in medical applications. Graph neural networks provide a powerful tool for analyzing the patients' features relative to each other. Recently, Graph Convolutional Networks (GCNs) have particularly been studied in the field of disease prediction. Due to the nature of such medical datasets, the class imbalance is a familiar issue in the field of disease prediction. When the class imbalance is present in the data, the existing graph-based classifiers tend to be biased towards the major class(es). Meanwhile, the correct diagnosis of the rare true-positive cases among all the patients is vital. In conventional methods, such imbalance is tackled by assigning appropriate weights to classes in the loss function; however, this solution is still dependent on the relative values of weights, sensitive to outliers, and in some cases biased towards the minor class(es). In this paper, we propose Re-weighted Adversarial Graph Convolutional Network (RA-GCN) to enhance the performance of the graph-based classifier and prevent it from emphasizing the samples of any particular class. This is accomplished by automatically learning to weigh the samples of the classes. For this purpose, a graph-based network is associated with each class, which is responsible for weighing the class samples and informing the classifier about the importance of each sample. Therefore, the classifier adjusts itself and determines the boundary between classes with more attention to the important samples. The parameters of the classifier and weighing networks are trained by an adversarial approach. At the end of the adversarial training process, the boundary of the classifier is more accurate and unbiased. We show the superiority of RA-GCN on synthetic and three publicly available medical datasets compared to the recent method.
Multi-layered Graph Embedding with Graph Convolutional Networks
Nov 22, 2018


Abstract:Recently, graph embedding emerges as an effective approach for graph analysis tasks such as node classification and link prediction. The goal of network embedding is to find low dimensional representation of graph nodes that preserves the graph structure. Since there might be signals on nodes as features, recent methods like Graph Convolutional Networks (GCNs) try to consider node signals besides the node relations. On the other hand, multi-layered graph analysis has been received much attention. However, the recent methods for node embedding have not been explored in these networks. In this paper, we study the problem of node embedding in multi-layered graphs and propose a deep method that embeds nodes using both relations (connections within and between layers of the graph) and nodes signals. We evaluate our method on node classification tasks. Experimental results demonstrate the superiority of the proposed method to other multi-layered and single-layered competitors and also proves the effect of using cross-layer edges.
Adversarial Classifier for Imbalanced Problems
Nov 21, 2018



Abstract:Adversarial approach has been widely used for data generation in the last few years. However, this approach has not been extensively utilized for classifier training. In this paper, we propose an adversarial framework for classifier training that can also handle imbalanced data. Indeed, a network is trained via an adversarial approach to give weights to samples of the majority class such that the obtained classification problem becomes more challenging for the discriminator and thus boosts its classification capability. In addition to the general imbalanced classification problems, the proposed method can also be used for problems such as graph representation learning in which it is desired to discriminate similar nodes from dissimilar nodes. Experimental results on imbalanced data classification and on the tasks like graph link prediction show the superiority of the proposed method compared to the state-of-the-art methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge