Teresa Tsang
Point Tracking as a Temporal Cue for Robust Myocardial Segmentation in Echocardiography Videos
Jan 14, 2026Abstract:Purpose: Myocardium segmentation in echocardiography videos is a challenging task due to low contrast, noise, and anatomical variability. Traditional deep learning models either process frames independently, ignoring temporal information, or rely on memory-based feature propagation, which accumulates error over time. Methods: We propose Point-Seg, a transformer-based segmentation framework that integrates point tracking as a temporal cue to ensure stable and consistent segmentation of myocardium across frames. Our method leverages a point-tracking module trained on a synthetic echocardiography dataset to track key anatomical landmarks across video sequences. These tracked trajectories provide an explicit motion-aware signal that guides segmentation, reducing drift and eliminating the need for memory-based feature accumulation. Additionally, we incorporate a temporal smoothing loss to further enhance temporal consistency across frames. Results: We evaluate our approach on both public and private echocardiography datasets. Experimental results demonstrate that Point-Seg has statistically similar accuracy in terms of Dice to state-of-the-art segmentation models in high quality echo data, while it achieves better segmentation accuracy in lower quality echo with improved temporal stability. Furthermore, Point-Seg has the key advantage of pixel-level myocardium motion information as opposed to other segmentation methods. Such information is essential in the computation of other downstream tasks such as myocardial strain measurement and regional wall motion abnormality detection. Conclusion: Point-Seg demonstrates that point tracking can serve as an effective temporal cue for consistent video segmentation, offering a reliable and generalizable approach for myocardium segmentation in echocardiography videos. The code is available at https://github.com/DeepRCL/PointSeg.
ControlEchoSynth: Boosting Ejection Fraction Estimation Models via Controlled Video Diffusion
Aug 25, 2025



Abstract:Synthetic data generation represents a significant advancement in boosting the performance of machine learning (ML) models, particularly in fields where data acquisition is challenging, such as echocardiography. The acquisition and labeling of echocardiograms (echo) for heart assessment, crucial in point-of-care ultrasound (POCUS) settings, often encounter limitations due to the restricted number of echo views available, typically captured by operators with varying levels of experience. This study proposes a novel approach for enhancing clinical diagnosis accuracy by synthetically generating echo views. These views are conditioned on existing, real views of the heart, focusing specifically on the estimation of ejection fraction (EF), a critical parameter traditionally measured from biplane apical views. By integrating a conditional generative model, we demonstrate an improvement in EF estimation accuracy, providing a comparative analysis with traditional methods. Preliminary results indicate that our synthetic echoes, when used to augment existing datasets, not only enhance EF estimation but also show potential in advancing the development of more robust, accurate, and clinically relevant ML models. This approach is anticipated to catalyze further research in synthetic data applications, paving the way for innovative solutions in medical imaging diagnostics.
Reliable Multi-View Learning with Conformal Prediction for Aortic Stenosis Classification in Echocardiography
Sep 15, 2024Abstract:The fundamental problem with ultrasound-guided diagnosis is that the acquired images are often 2-D cross-sections of a 3-D anatomy, potentially missing important anatomical details. This limitation leads to challenges in ultrasound echocardiography, such as poor visualization of heart valves or foreshortening of ventricles. Clinicians must interpret these images with inherent uncertainty, a nuance absent in machine learning's one-hot labels. We propose Re-Training for Uncertainty (RT4U), a data-centric method to introduce uncertainty to weakly informative inputs in the training set. This simple approach can be incorporated to existing state-of-the-art aortic stenosis classification methods to further improve their accuracy. When combined with conformal prediction techniques, RT4U can yield adaptively sized prediction sets which are guaranteed to contain the ground truth class to a high accuracy. We validate the effectiveness of RT4U on three diverse datasets: a public (TMED-2) and a private AS dataset, along with a CIFAR-10-derived toy dataset. Results show improvement on all the datasets.
CCSI: Continual Class-Specific Impression for Data-free Class Incremental Learning
Jun 09, 2024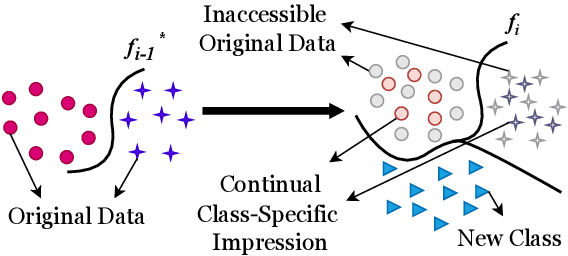

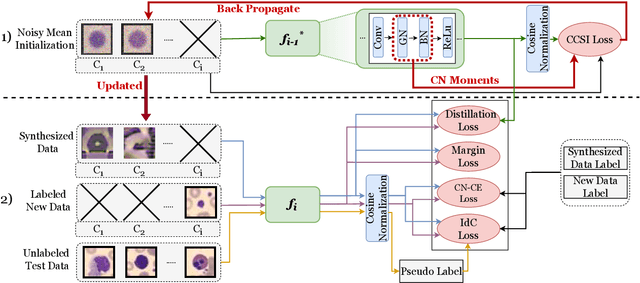
Abstract:In real-world clinical settings, traditional deep learning-based classification methods struggle with diagnosing newly introduced disease types because they require samples from all disease classes for offline training. Class incremental learning offers a promising solution by adapting a deep network trained on specific disease classes to handle new diseases. However, catastrophic forgetting occurs, decreasing the performance of earlier classes when adapting the model to new data. Prior proposed methodologies to overcome this require perpetual storage of previous samples, posing potential practical concerns regarding privacy and storage regulations in healthcare. To this end, we propose a novel data-free class incremental learning framework that utilizes data synthesis on learned classes instead of data storage from previous classes. Our key contributions include acquiring synthetic data known as Continual Class-Specific Impression (CCSI) for previously inaccessible trained classes and presenting a methodology to effectively utilize this data for updating networks when introducing new classes. We obtain CCSI by employing data inversion over gradients of the trained classification model on previous classes starting from the mean image of each class inspired by common landmarks shared among medical images and utilizing continual normalization layers statistics as a regularizer in this pixel-wise optimization process. Subsequently, we update the network by combining the synthesized data with new class data and incorporate several losses, including an intra-domain contrastive loss to generalize the deep network trained on the synthesized data to real data, a margin loss to increase separation among previous classes and new ones, and a cosine-normalized cross-entropy loss to alleviate the adverse effects of imbalanced distributions in training data.
EchoGNN: Explainable Ejection Fraction Estimation with Graph Neural Networks
Aug 30, 2022



Abstract:Ejection fraction (EF) is a key indicator of cardiac function, allowing identification of patients prone to heart dysfunctions such as heart failure. EF is estimated from cardiac ultrasound videos known as echocardiograms (echo) by manually tracing the left ventricle and estimating its volume on certain frames. These estimations exhibit high inter-observer variability due to the manual process and varying video quality. Such sources of inaccuracy and the need for rapid assessment necessitate reliable and explainable machine learning techniques. In this work, we introduce EchoGNN, a model based on graph neural networks (GNNs) to estimate EF from echo videos. Our model first infers a latent echo-graph from the frames of one or multiple echo cine series. It then estimates weights over nodes and edges of this graph, indicating the importance of individual frames that aid EF estimation. A GNN regressor uses this weighted graph to predict EF. We show, qualitatively and quantitatively, that the learned graph weights provide explainability through identification of critical frames for EF estimation, which can be used to determine when human intervention is required. On EchoNet-Dynamic public EF dataset, EchoGNN achieves EF prediction performance that is on par with state of the art and provides explainability, which is crucial given the high inter-observer variability inherent in this task.
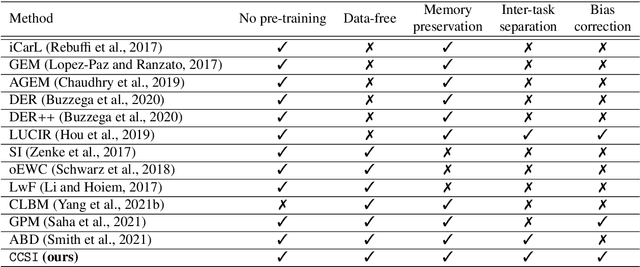
Class Impression for Data-free Incremental Learning
Jul 04, 2022
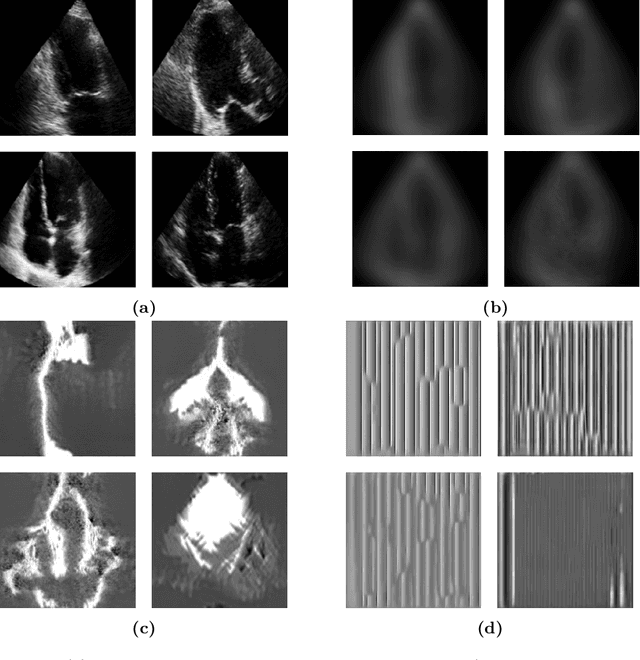
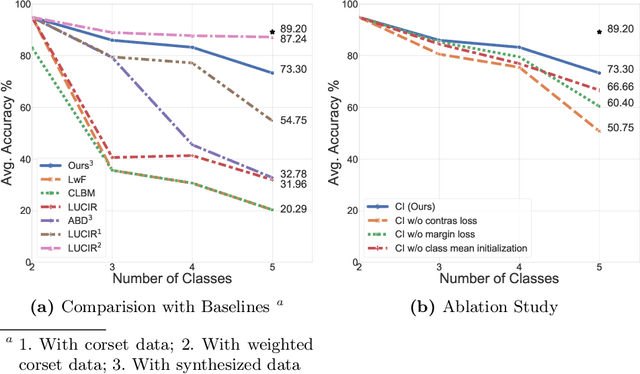
Abstract:Standard deep learning-based classification approaches require collecting all samples from all classes in advance and are trained offline. This paradigm may not be practical in real-world clinical applications, where new classes are incrementally introduced through the addition of new data. Class incremental learning is a strategy allowing learning from such data. However, a major challenge is catastrophic forgetting, i.e., performance degradation on previous classes when adapting a trained model to new data. Prior methodologies to alleviate this challenge save a portion of training data require perpetual storage of such data that may introduce privacy issues. Here, we propose a novel data-free class incremental learning framework that first synthesizes data from the model trained on previous classes to generate a \ours. Subsequently, it updates the model by combining the synthesized data with new class data. Furthermore, we incorporate a cosine normalized Cross-entropy loss to mitigate the adverse effects of the imbalance, a margin loss to increase separation among previous classes and new ones, and an intra-domain contrastive loss to generalize the model trained on the synthesized data to real data. We compare our proposed framework with state-of-the-art methods in class incremental learning, where we demonstrate improvement in accuracy for the classification of 11,062 echocardiography cine series of patients.
Echo-SyncNet: Self-supervised Cardiac View Synchronization in Echocardiography
Feb 03, 2021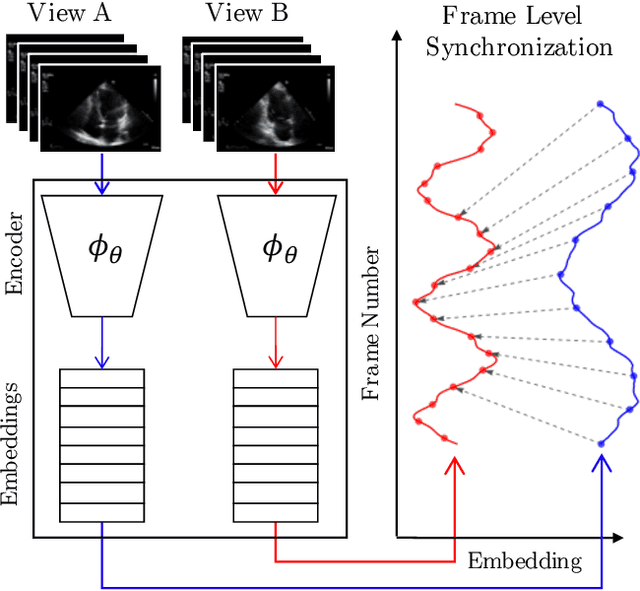
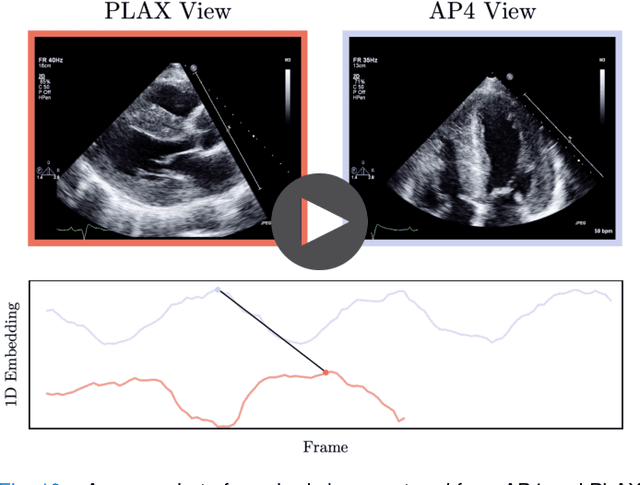
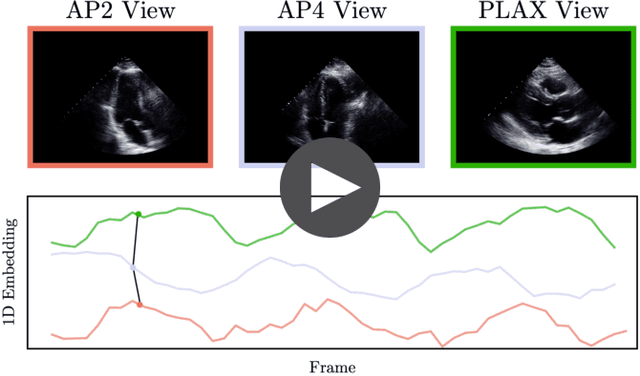
Abstract:In echocardiography (echo), an electrocardiogram (ECG) is conventionally used to temporally align different cardiac views for assessing critical measurements. However, in emergencies or point-of-care situations, acquiring an ECG is often not an option, hence motivating the need for alternative temporal synchronization methods. Here, we propose Echo-SyncNet, a self-supervised learning framework to synchronize various cross-sectional 2D echo series without any external input. The proposed framework takes advantage of both intra-view and inter-view self supervisions. The former relies on spatiotemporal patterns found between the frames of a single echo cine and the latter on the interdependencies between multiple cines. The combined supervisions are used to learn a feature-rich embedding space where multiple echo cines can be temporally synchronized. We evaluate the framework with multiple experiments: 1) Using data from 998 patients, Echo-SyncNet shows promising results for synchronizing Apical 2 chamber and Apical 4 chamber cardiac views; 2) Using data from 3070 patients, our experiments reveal that the learned representations of Echo-SyncNet outperform a supervised deep learning method that is optimized for automatic detection of fine-grained cardiac phase; 3) We show the usefulness of the learned representations in a one-shot learning scenario of cardiac keyframe detection. Without any fine-tuning, keyframes in 1188 validation patient studies are identified by synchronizing them with only one labeled reference study. We do not make any prior assumption about what specific cardiac views are used for training and show that Echo-SyncNet can accurately generalize to views not present in its training set. Project repository: github.com/fatemehtd/Echo-SyncNet.
U-LanD: Uncertainty-Driven Video Landmark Detection
Feb 02, 2021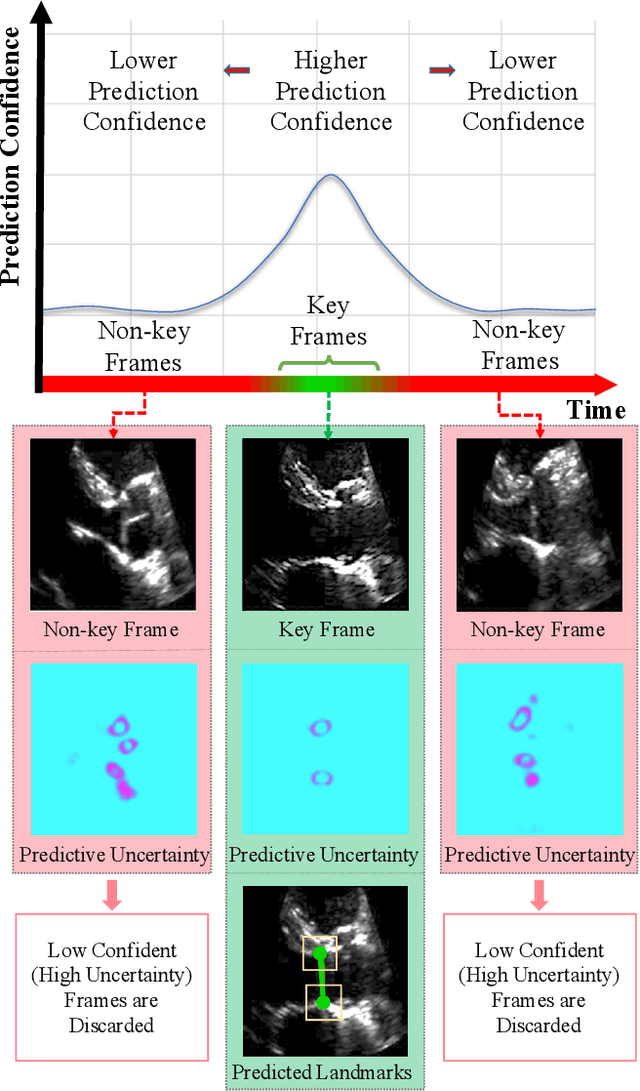
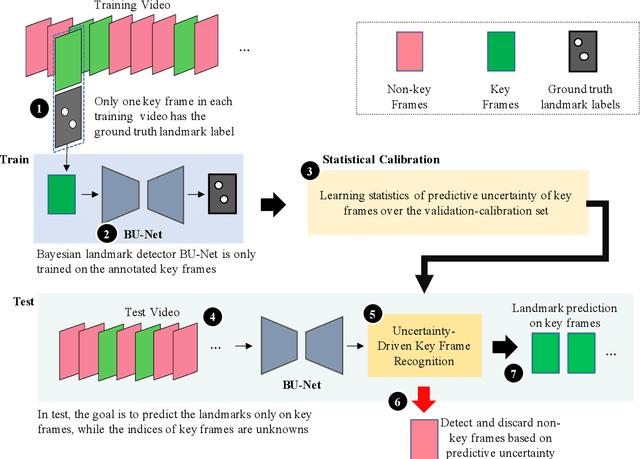
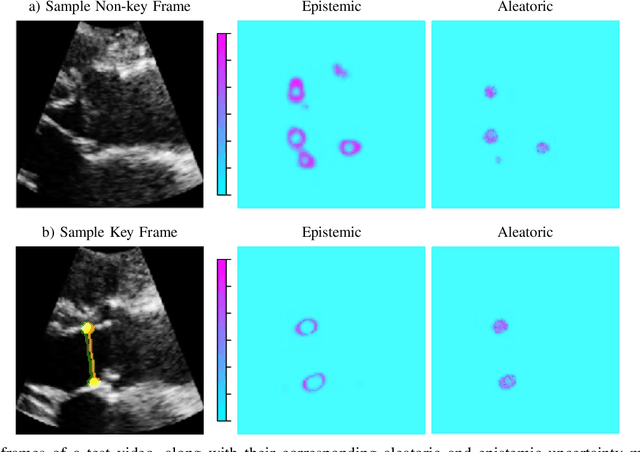
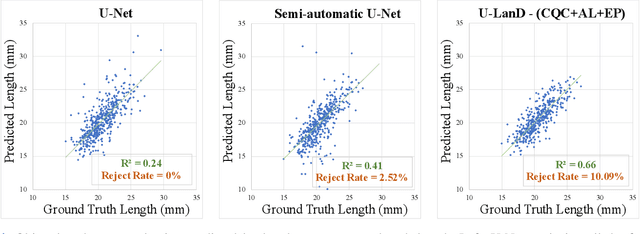
Abstract:This paper presents U-LanD, a framework for joint detection of key frames and landmarks in videos. We tackle a specifically challenging problem, where training labels are noisy and highly sparse. U-LanD builds upon a pivotal observation: a deep Bayesian landmark detector solely trained on key video frames, has significantly lower predictive uncertainty on those frames vs. other frames in videos. We use this observation as an unsupervised signal to automatically recognize key frames on which we detect landmarks. As a test-bed for our framework, we use ultrasound imaging videos of the heart, where sparse and noisy clinical labels are only available for a single frame in each video. Using data from 4,493 patients, we demonstrate that U-LanD can exceedingly outperform the state-of-the-art non-Bayesian counterpart by a noticeable absolute margin of 42% in R2 score, with almost no overhead imposed on the model size. Our approach is generic and can be potentially applied to other challenging data with noisy and sparse training labels.
Reciprocal Landmark Detection and Tracking with Extremely Few Annotations
Jan 27, 2021



Abstract:Localization of anatomical landmarks to perform two-dimensional measurements in echocardiography is part of routine clinical workflow in cardiac disease diagnosis. Automatic localization of those landmarks is highly desirable to improve workflow and reduce interobserver variability. Training a machine learning framework to perform such localization is hindered given the sparse nature of gold standard labels; only few percent of cardiac cine series frames are normally manually labeled for clinical use. In this paper, we propose a new end-to-end reciprocal detection and tracking model that is specifically designed to handle the sparse nature of echocardiography labels. The model is trained using few annotated frames across the entire cardiac cine sequence to generate consistent detection and tracking of landmarks, and an adversarial training for the model is proposed to take advantage of these annotated frames. The superiority of the proposed reciprocal model is demonstrated using a series of experiments.
GAN-enhanced Conditional Echocardiogram Generation
Nov 23, 2019


Abstract:Echocardiography (echo) is a common means of evaluating cardiac conditions. Due to the label scarcity, semi-supervised paradigms in automated echo analysis are getting traction. One of the most sought-after problems in echo is the segmentation of cardiac structures (e.g. chambers). Accordingly, we propose an echocardiogram generation approach using generative adversarial networks with a conditional patch-based discriminator. In this work, we validate the feasibility of GAN-enhanced echo generation with different conditions (segmentation masks), namely, the left ventricle, ventricular myocardium, and atrium. Results show that the proposed adversarial algorithm can generate high-quality echo frames whose cardiac structures match the given segmentation masks. This method is expected to facilitate the training of other machine learning models in a semi-supervised fashion as suggested in similar researches.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge