Robert Rohling
ResNet Structure Simplification with the Convolutional Kernel Redundancy Measure
Dec 01, 2022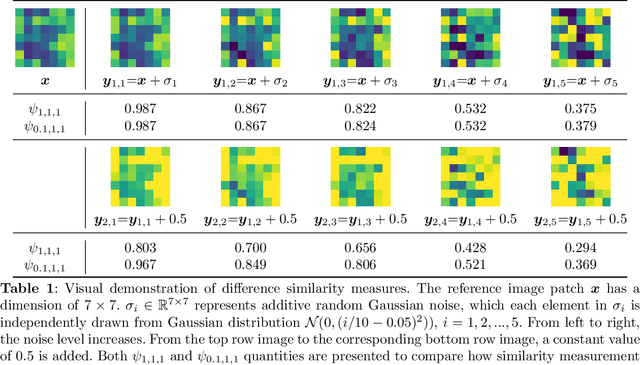
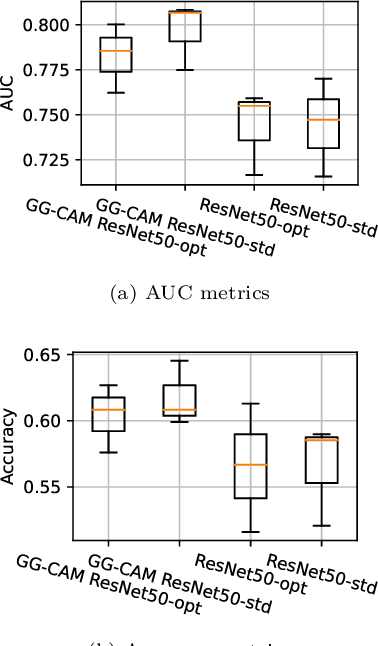
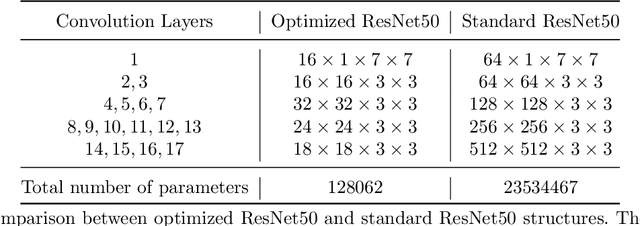
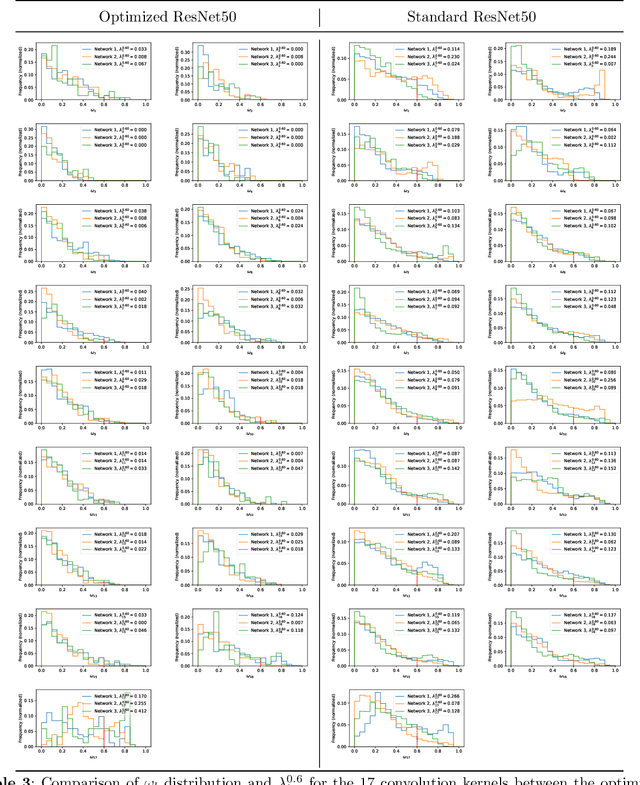
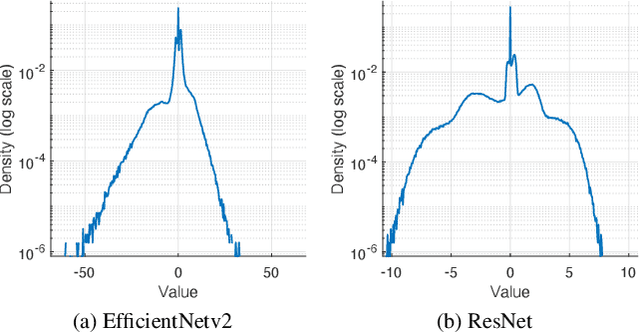
Abstract:Deep learning, especially convolutional neural networks, has triggered accelerated advancements in computer vision, bringing changes into our daily practice. Furthermore, the standardized deep learning modules (also known as backbone networks), i.e., ResNet and EfficientNet, have enabled efficient and rapid development of new computer vision solutions. Yet, deep learning methods still suffer from several drawbacks. One of the most concerning problems is the high memory and computational cost, such that dedicated computing units, typically GPUs, have to be used for training and development. Therefore, in this paper, we propose a quantifiable evaluation method, the convolutional kernel redundancy measure, which is based on perceived image differences, for guiding the network structure simplification. When applying our method to the chest X-ray image classification problem with ResNet, our method can maintain the performance of the network and reduce the number of parameters from over $23$ million to approximately $128$ thousand (reducing $99.46\%$ of the parameters).
The Open Kidney Ultrasound Data Set
Jun 14, 2022



Abstract:Ultrasound use is because of its low cost, non-ionizing, and non-invasive characteristics, and has established itself as a cornerstone radiological examination. Research on ultrasound applications has also expanded, especially with image analysis with machine learning. However, ultrasound data are frequently restricted to closed data sets, with only a few openly available. Despite being a frequently examined organ, the kidney lacks a publicly available ultrasonography data set. The proposed Open Kidney Ultrasound Data Set is the first publicly available set of kidney B-mode ultrasound data that includes annotations for multi-class semantic segmentation. It is based on data retrospectively collected in a 5-year period from over 500 patients with a mean age of 53.2 +/- 14.7 years, body mass index of 27.0 +/- 5.4 kg/m2, and most common primary diseases being diabetes mellitus, IgA nephropathy, and hypertension. There are labels for the view and fine-grained manual annotations from two expert sonographers. Notably, this data includes native and transplanted kidneys. Initial benchmarking measurements are performed, demonstrating a state-of-the-art algorithm achieving a Dice Sorenson Coefficient of 0.74 for the kidney capsule. This data set is a high-quality data set, including two sets of expert annotations, with a larger breadth of images than previously available. In increasing access to kidney ultrasound data, future researchers may be able to create novel image analysis techniques for tissue characterization, disease detection, and prognostication.
The Kidneys Are Not All Normal: Investigating the Speckle Distributions of Transplanted Kidneys
Jun 14, 2022


Abstract:Modelling ultrasound speckle has generated considerable interest for its ability to characterize tissue properties. As speckle is dependent on the underlying tissue architecture, modelling it may aid in tasks like segmentation or disease detection. However, for the transplanted kidney where ultrasound is commonly used to investigate dysfunction, it is currently unknown which statistical distribution best characterises such speckle. This is especially true for the regions of the transplanted kidney: the cortex, the medulla and the central echogenic complex. Furthermore, it is unclear how these distributions vary by patient variables such as age, sex, body mass index, primary disease, or donor type. These traits may influence speckle modelling given their influence on kidney anatomy. We are the first to investigate these two aims. N=821 kidney transplant recipient B-mode images were automatically segmented into the cortex, medulla, and central echogenic complex using a neural network. Seven distinct probability distributions were fitted to each region. The Rayleigh and Nakagami distributions had model parameters that differed significantly between the three regions (p <= 0.05). While both had excellent goodness of fit, the Nakagami had higher Kullbeck-Leibler divergence. Recipient age correlated weakly with scale in the cortex (Omega: rho = 0.11, p = 0.004), while body mass index correlated weakly with shape in the medulla (m: rho = 0.08, p = 0.04). Neither sex, primary disease, nor donor type demonstrated any correlation. We propose the Nakagami distribution be used to characterize transplanted kidneys regionally independent of disease etiology and most patient characteristics based on our findings.
Quasi-Real Time Multi-Frequency 3D Shear Wave Absolute Vibro-Elastography System for Prostate
May 09, 2022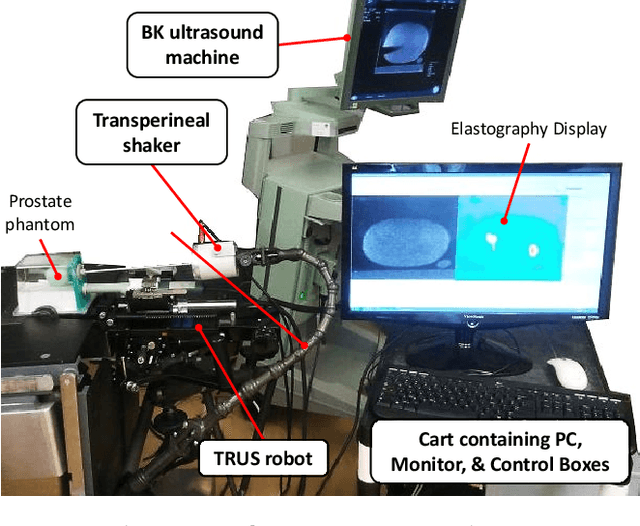
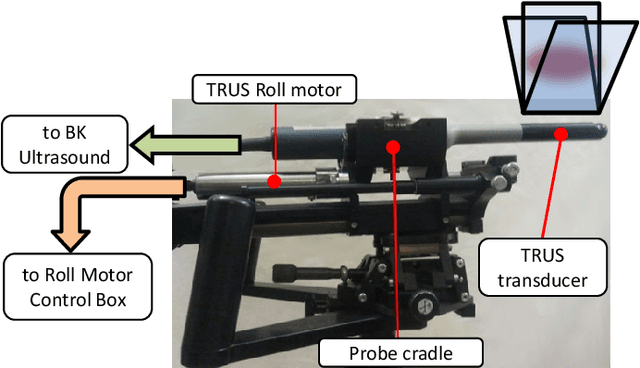
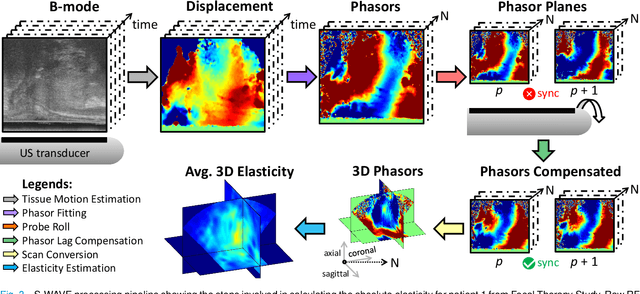
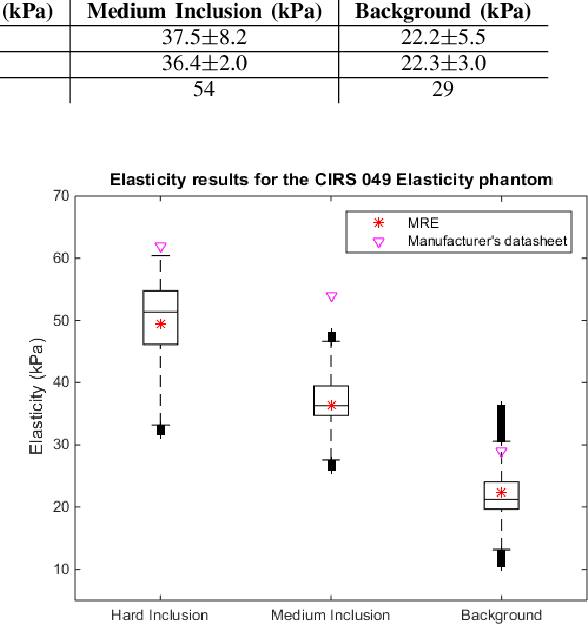
Abstract:This article describes a novel quasi-real time system for quantitative and volumetric measurement of tissue elasticity in the prostate. Tissue elasticity is computed by using a local frequency estimator to measure the three dimensional local wavelengths of a steady-state shear wave within the prostate gland. The shear wave is created using a mechanical voice coil shaker which transmits multi-frequency vibrations transperineally. Radio frequency data is streamed directly from a BK Medical 8848 trans-rectal ultrasound transducer to an external computer where tissue displacement due to the excitation is measured using a speckle tracking algorithm. Bandpass sampling is used that eliminates the need for an ultra fast frame rate to track the tissue motion and allows for accurate reconstruction at a sampling frequency that is below the Nyquist rate. A roll motor with computer control is used to rotate the sagittal array of the transducer and obtain the 3D data. Two CIRS phantoms were used to validate both the accuracy of the elasticity measurement as well as the functional feasibility of using the system for in vivo prostate imaging. The system has been used in two separate clinical studies as a method for cancer identification. The results, presented here, show numerical and visual correlations between our stiffness measurements and cancer likelihood as determined from pathology results. Initial published results using this system include an area under the receiver operating characteristic curve of 0.82+/-0.01 with regards to prostate cancer identification in the peripheral zone.
Multi-task UNet: Jointly Boosting Saliency Prediction and Disease Classification on Chest X-ray Images
Feb 15, 2022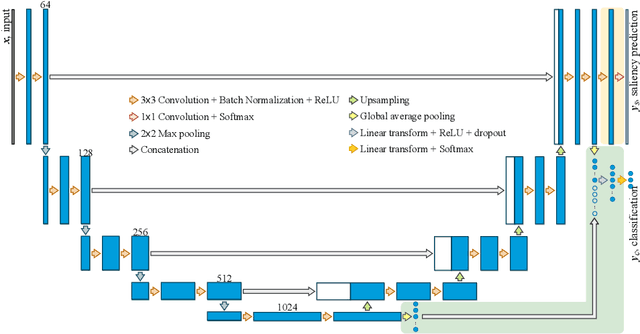
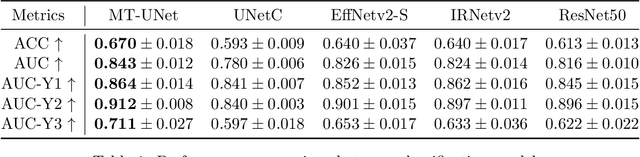


Abstract:Human visual attention has recently shown its distinct capability in boosting machine learning models. However, studies that aim to facilitate medical tasks with human visual attention are still scarce. To support the use of visual attention, this paper describes a novel deep learning model for visual saliency prediction on chest X-ray (CXR) images. To cope with data deficiency, we exploit the multi-task learning method and tackles disease classification on CXR simultaneously. For a more robust training process, we propose a further optimized multi-task learning scheme to better handle model overfitting. Experiments show our proposed deep learning model with our new learning scheme can outperform existing methods dedicated either for saliency prediction or image classification. The code used in this paper is available at https://github.com/hz-zhu/MT-UNet.
Gaze-Guided Class Activation Mapping: Leveraging Human Attention for Network Attention in Chest X-rays Classification
Feb 15, 2022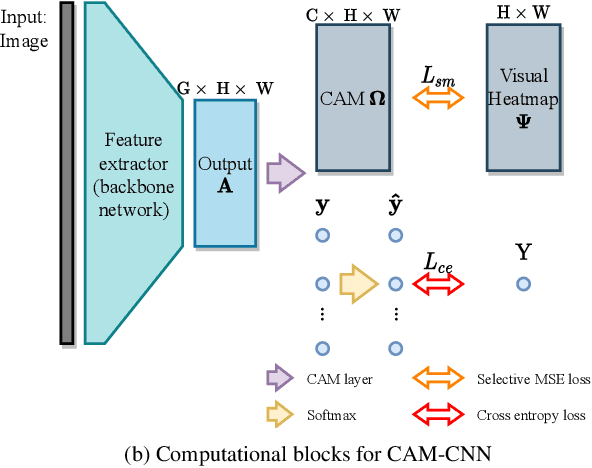
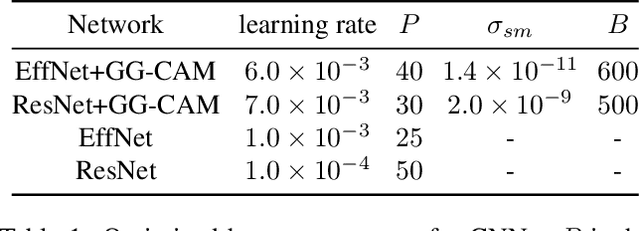
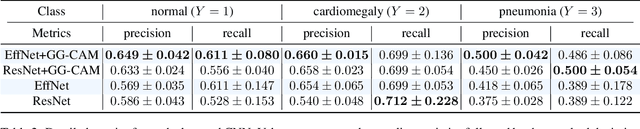
Abstract:The increased availability and accuracy of eye-gaze tracking technology has sparked attention-related research in psychology, neuroscience, and, more recently, computer vision and artificial intelligence. The attention mechanism in artificial neural networks is known to improve learning tasks. However, no previous research has combined the network attention and human attention. This paper describes a gaze-guided class activation mapping (GG-CAM) method to directly regulate the formation of network attention based on expert radiologists' visual attention for the chest X-ray pathology classification problem, which remains challenging due to the complex and often nuanced differences among images. GG-CAM is a lightweight ($3$ additional trainable parameters for regulating the learning process) and generic extension that can be easily applied to most classification convolutional neural networks (CNN). GG-CAM-modified CNNs do not require human attention as an input when fully trained. Comparative experiments suggest that two standard CNNs with the GG-CAM extension achieve significantly greater classification performance. The median area under the curve (AUC) metrics for ResNet50 increases from $0.721$ to $0.776$. For EfficientNetv2 (s), the median AUC increases from $0.723$ to $0.801$. The GG-CAM also brings better interpretability of the network that facilitates the weakly-supervised pathology localization and analysis.
Multifrequency 3D Elasticity Reconstruction withStructured Sparsity and ADMM
Nov 23, 2021
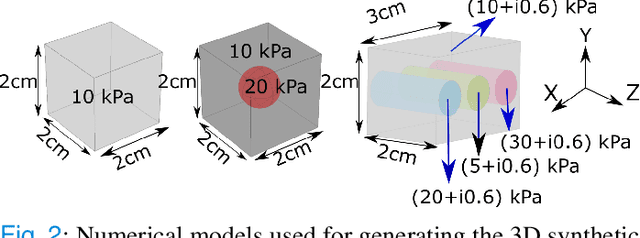

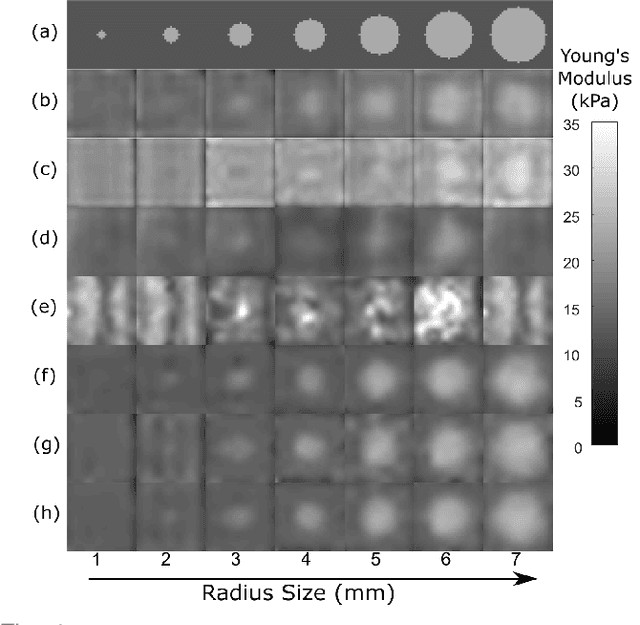
Abstract:We introduce a model-based iterative method to obtain shear modulus images of tissue using magnetic resonance elastography. The method jointly finds the displacement field that best fits multifrequency tissue displacement data and the corresponding shear modulus. The displacement satisfies a viscoelastic wave equation constraint, discretized using the finite element method. Sparsifying regularization terms in both shear modulus and the displacement are used in the cost function minimized for the best fit. The formulated problem is bi-convex. Its solution can be obtained iteratively by using the alternating direction method of multipliers. Sparsifying regularizations and the wave equation constraint filter out sensor noise and compressional waves. Our method does not require bandpass filtering as a preprocessing step and converges fast irrespective of the initialization. We evaluate our new method in multiple in silico and phantom experiments, with comparisons with existing methods, and we show improvements in contrast to noise and signal to noise ratios. Results from an in vivo liver imaging study show elastograms with mean elasticity comparable to other values reported in the literature.
Echo-SyncNet: Self-supervised Cardiac View Synchronization in Echocardiography
Feb 03, 2021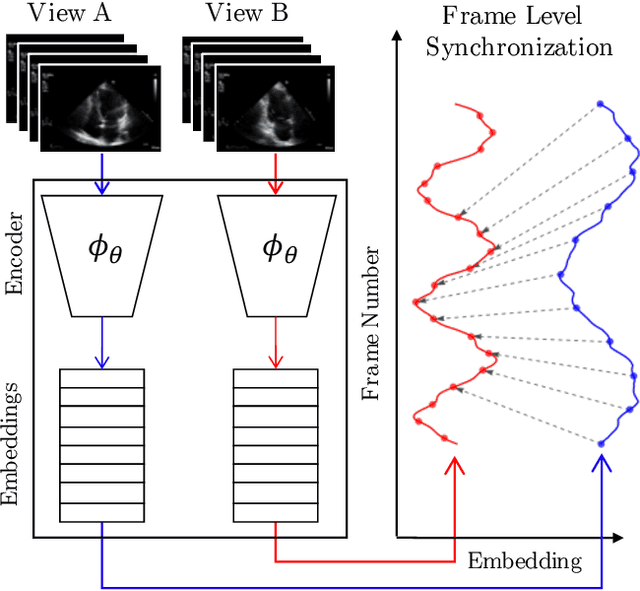
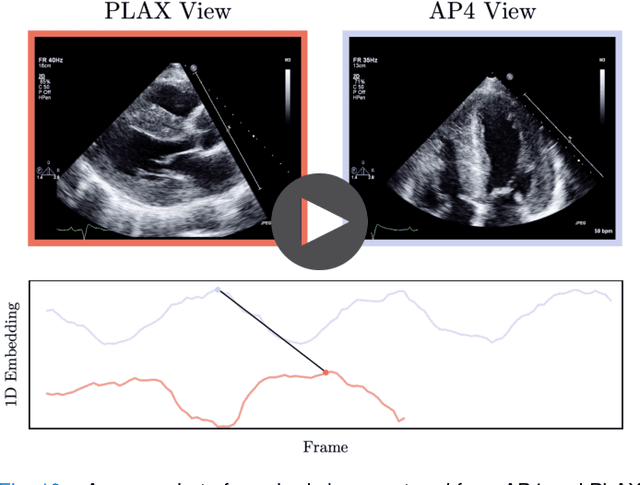
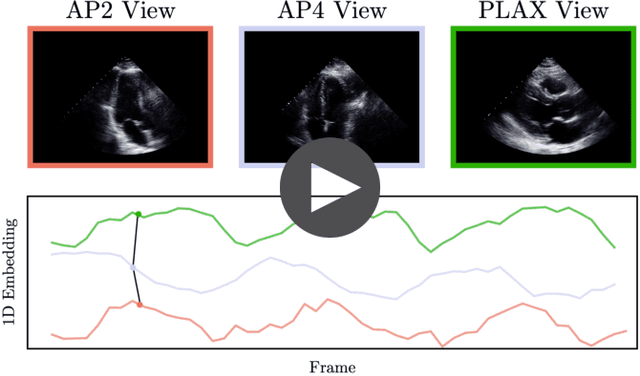
Abstract:In echocardiography (echo), an electrocardiogram (ECG) is conventionally used to temporally align different cardiac views for assessing critical measurements. However, in emergencies or point-of-care situations, acquiring an ECG is often not an option, hence motivating the need for alternative temporal synchronization methods. Here, we propose Echo-SyncNet, a self-supervised learning framework to synchronize various cross-sectional 2D echo series without any external input. The proposed framework takes advantage of both intra-view and inter-view self supervisions. The former relies on spatiotemporal patterns found between the frames of a single echo cine and the latter on the interdependencies between multiple cines. The combined supervisions are used to learn a feature-rich embedding space where multiple echo cines can be temporally synchronized. We evaluate the framework with multiple experiments: 1) Using data from 998 patients, Echo-SyncNet shows promising results for synchronizing Apical 2 chamber and Apical 4 chamber cardiac views; 2) Using data from 3070 patients, our experiments reveal that the learned representations of Echo-SyncNet outperform a supervised deep learning method that is optimized for automatic detection of fine-grained cardiac phase; 3) We show the usefulness of the learned representations in a one-shot learning scenario of cardiac keyframe detection. Without any fine-tuning, keyframes in 1188 validation patient studies are identified by synchronizing them with only one labeled reference study. We do not make any prior assumption about what specific cardiac views are used for training and show that Echo-SyncNet can accurately generalize to views not present in its training set. Project repository: github.com/fatemehtd/Echo-SyncNet.
U-LanD: Uncertainty-Driven Video Landmark Detection
Feb 02, 2021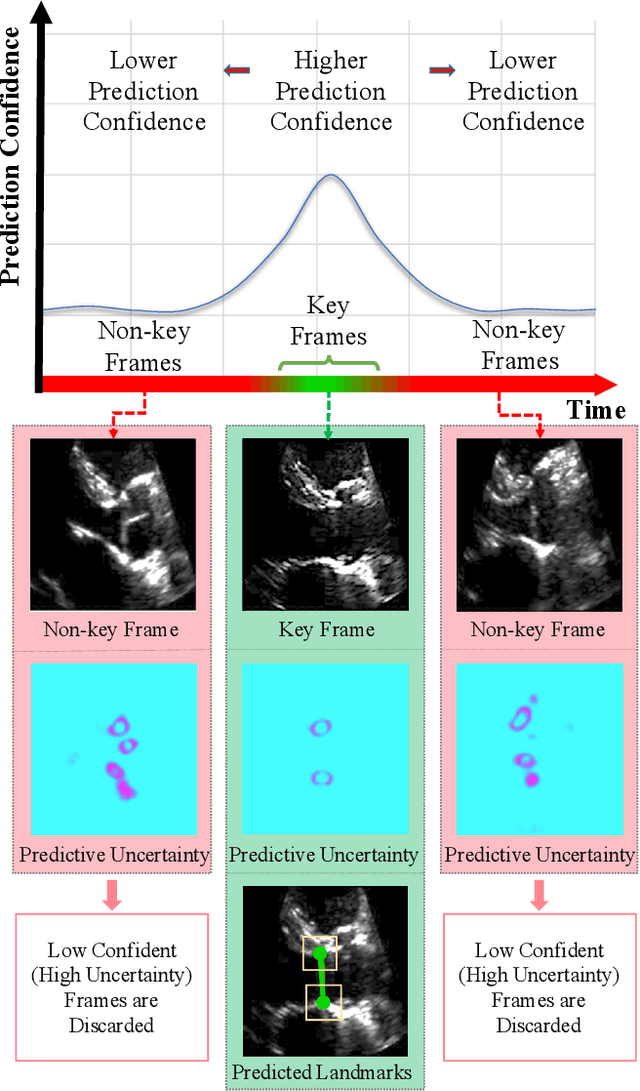
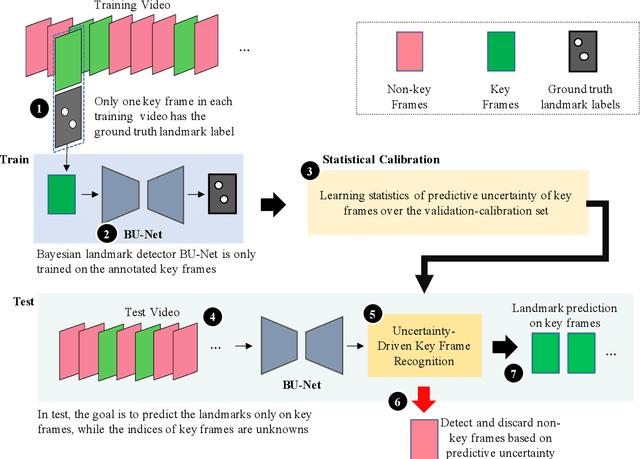
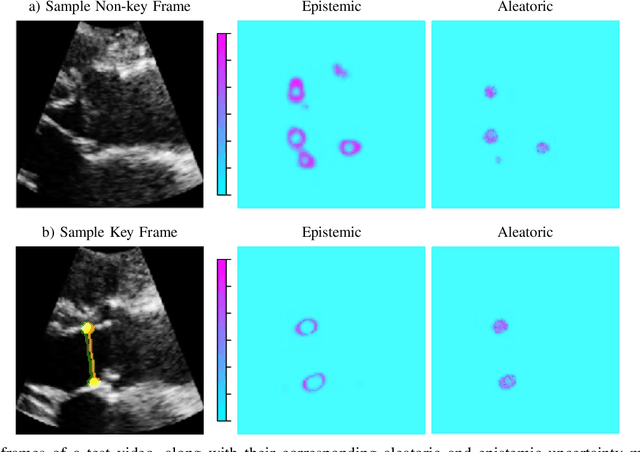
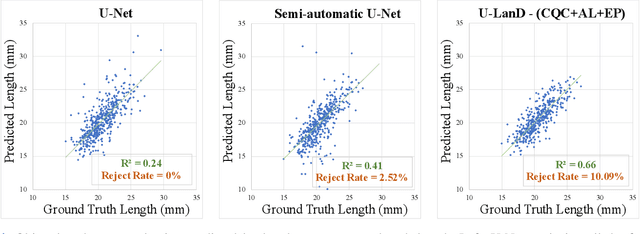
Abstract:This paper presents U-LanD, a framework for joint detection of key frames and landmarks in videos. We tackle a specifically challenging problem, where training labels are noisy and highly sparse. U-LanD builds upon a pivotal observation: a deep Bayesian landmark detector solely trained on key video frames, has significantly lower predictive uncertainty on those frames vs. other frames in videos. We use this observation as an unsupervised signal to automatically recognize key frames on which we detect landmarks. As a test-bed for our framework, we use ultrasound imaging videos of the heart, where sparse and noisy clinical labels are only available for a single frame in each video. Using data from 4,493 patients, we demonstrate that U-LanD can exceedingly outperform the state-of-the-art non-Bayesian counterpart by a noticeable absolute margin of 42% in R2 score, with almost no overhead imposed on the model size. Our approach is generic and can be potentially applied to other challenging data with noisy and sparse training labels.
On Modelling Label Uncertainty in Deep Neural Networks: Automatic Estimation of Intra-observer Variability in 2D Echocardiography Quality Assessment
Nov 02, 2019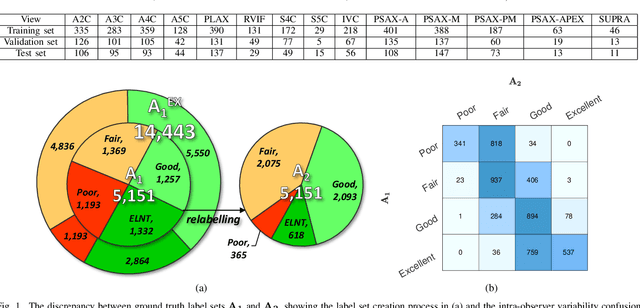
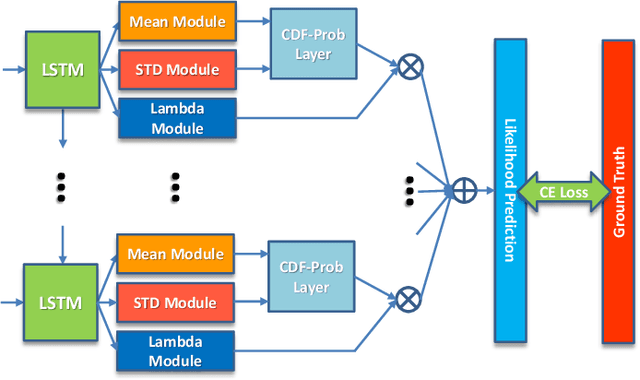
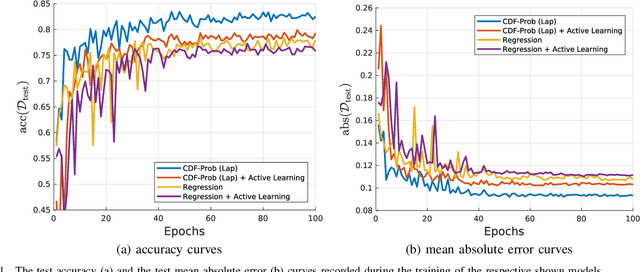
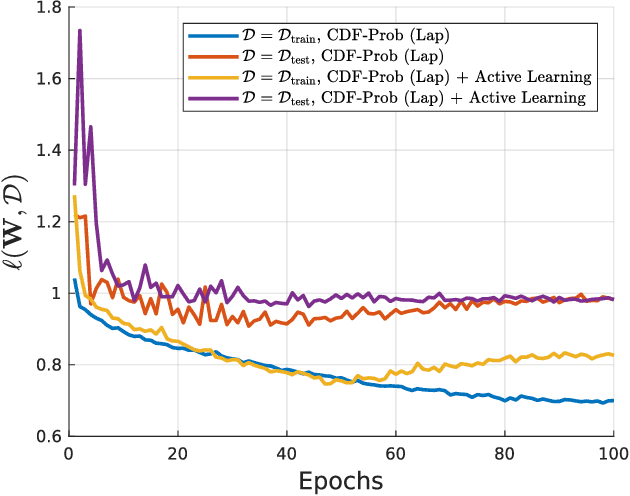
Abstract:Uncertainty of labels in clinical data resulting from intra-observer variability can have direct impact on the reliability of assessments made by deep neural networks. In this paper, we propose a method for modelling such uncertainty in the context of 2D echocardiography (echo), which is a routine procedure for detecting cardiovascular disease at point-of-care. Echo imaging quality and acquisition time is highly dependent on the operator's experience level. Recent developments have shown the possibility of automating echo image quality quantification by mapping an expert's assessment of quality to the echo image via deep learning techniques. Nevertheless, the observer variability in the expert's assessment can impact the quality quantification accuracy. Here, we aim to model the intra-observer variability in echo quality assessment as an aleatoric uncertainty modelling regression problem with the introduction of a novel method that handles the regression problem with categorical labels. A key feature of our design is that only a single forward pass is sufficient to estimate the level of uncertainty for the network output. Compared to the $0.11 \pm 0.09$ absolute error (in a scale from 0 to 1) archived by the conventional regression method, the proposed method brings the error down to $0.09 \pm 0.08$, where the improvement is statistically significant and equivalents to $5.7\%$ test accuracy improvement. The simplicity of the proposed approach means that it could be generalized to other applications of deep learning in medical imaging, where there is often uncertainty in clinical labels.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge