William Wells
Unified Cross-Modal Image Synthesis with Hierarchical Mixture of Product-of-Experts
Oct 25, 2024Abstract:We propose a deep mixture of multimodal hierarchical variational auto-encoders called MMHVAE that synthesizes missing images from observed images in different modalities. MMHVAE's design focuses on tackling four challenges: (i) creating a complex latent representation of multimodal data to generate high-resolution images; (ii) encouraging the variational distributions to estimate the missing information needed for cross-modal image synthesis; (iii) learning to fuse multimodal information in the context of missing data; (iv) leveraging dataset-level information to handle incomplete data sets at training time. Extensive experiments are performed on the challenging problem of pre-operative brain multi-parametric magnetic resonance and intra-operative ultrasound imaging.
Patient-Specific Real-Time Segmentation in Trackerless Brain Ultrasound
May 16, 2024



Abstract:Intraoperative ultrasound (iUS) imaging has the potential to improve surgical outcomes in brain surgery. However, its interpretation is challenging, even for expert neurosurgeons. In this work, we designed the first patient-specific framework that performs brain tumor segmentation in trackerless iUS. To disambiguate ultrasound imaging and adapt to the neurosurgeon's surgical objective, a patient-specific real-time network is trained using synthetic ultrasound data generated by simulating virtual iUS sweep acquisitions in pre-operative MR data. Extensive experiments performed in real ultrasound data demonstrate the effectiveness of the proposed approach, allowing for adapting to the surgeon's definition of surgical targets and outperforming non-patient-specific models, neurosurgeon experts, and high-end tracking systems. Our code is available at: \url{https://github.com/ReubenDo/MHVAE-Seg}.
Unimodal Cyclic Regularization for Training Multimodal Image Registration Networks
Nov 12, 2020



Abstract:The loss function of an unsupervised multimodal image registration framework has two terms, i.e., a metric for similarity measure and regularization. In the deep learning era, researchers proposed many approaches to automatically learn the similarity metric, which has been shown effective in improving registration performance. However, for the regularization term, most existing multimodal registration approaches still use a hand-crafted formula to impose artificial properties on the estimated deformation field. In this work, we propose a unimodal cyclic regularization training pipeline, which learns task-specific prior knowledge from simpler unimodal registration, to constrain the deformation field of multimodal registration. In the experiment of abdominal CT-MR registration, the proposed method yields better results over conventional regularization methods, especially for severely deformed local regions.
Joint Modeling of Chest Radiographs and Radiology Reports for Pulmonary Edema Assessment
Aug 22, 2020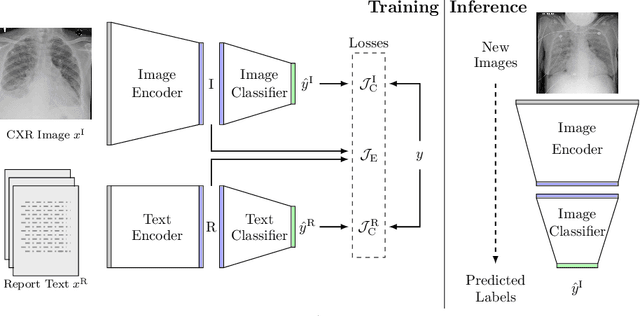
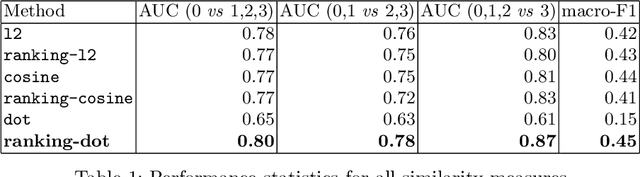

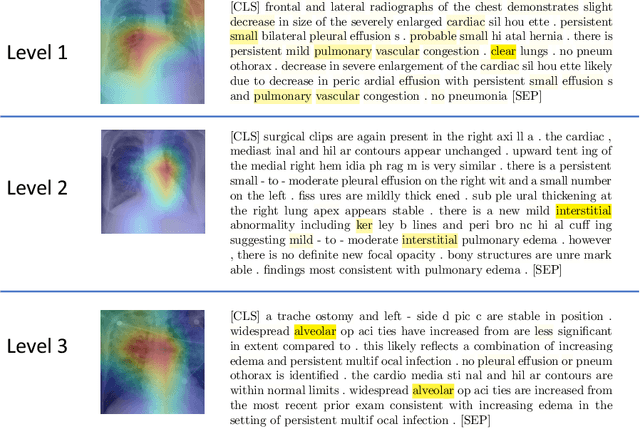
Abstract:We propose and demonstrate a novel machine learning algorithm that assesses pulmonary edema severity from chest radiographs. While large publicly available datasets of chest radiographs and free-text radiology reports exist, only limited numerical edema severity labels can be extracted from radiology reports. This is a significant challenge in learning such models for image classification. To take advantage of the rich information present in the radiology reports, we develop a neural network model that is trained on both images and free-text to assess pulmonary edema severity from chest radiographs at inference time. Our experimental results suggest that the joint image-text representation learning improves the performance of pulmonary edema assessment compared to a supervised model trained on images only. We also show the use of the text for explaining the image classification by the joint model. To the best of our knowledge, our approach is the first to leverage free-text radiology reports for improving the image model performance in this application. Our code is available at https://github.com/RayRuizhiLiao/joint_chestxray.
Semi-supervised Learning for Quantification of Pulmonary Edema in Chest X-Ray Images
Apr 10, 2019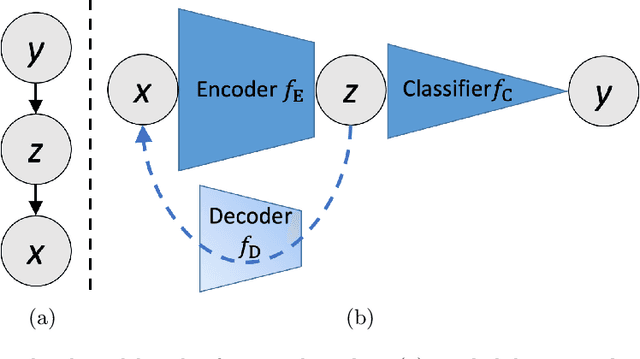
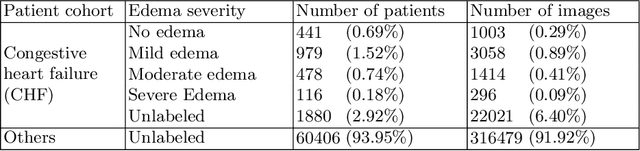
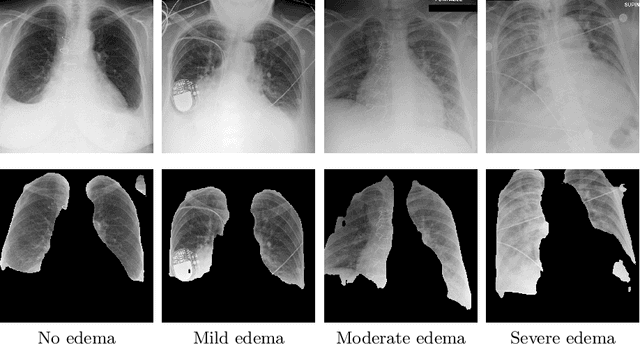
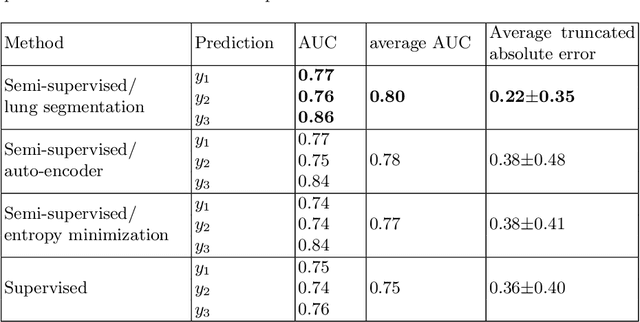
Abstract:We propose and demonstrate machine learning algorithms to assess the severity of pulmonary edema in chest x-ray images of congestive heart failure patients. Accurate assessment of pulmonary edema in heart failure is critical when making treatment and disposition decisions. Our work is grounded in a large-scale clinical dataset of over 300,000 x-ray images with associated radiology reports. While edema severity labels can be extracted unambiguously from a small fraction of the radiology reports, accurate annotation is challenging in most cases. To take advantage of the unlabeled images, we develop a Bayesian model that includes a variational auto-encoder for learning a latent representation from the entire image set trained jointly with a regressor that employs this representation for predicting pulmonary edema severity. Our experimental results suggest that modeling the distribution of images jointly with the limited labels improves the accuracy of pulmonary edema scoring compared to a strictly supervised approach. To the best of our knowledge, this is the first attempt to employ machine learning algorithms to automatically and quantitatively assess the severity of pulmonary edema in chest x-ray images.
Keypoint Transfer for Fast Whole-Body Segmentation
Jun 22, 2018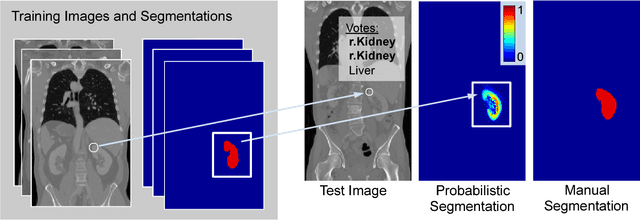
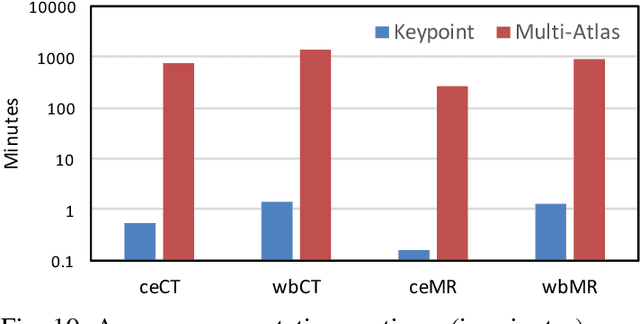
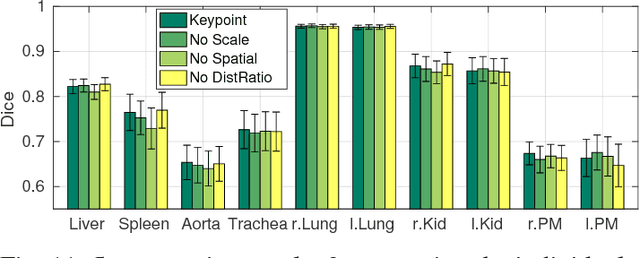
Abstract:We introduce an approach for image segmentation based on sparse correspondences between keypoints in testing and training images. Keypoints represent automatically identified distinctive image locations, where each keypoint correspondence suggests a transformation between images. We use these correspondences to transfer label maps of entire organs from the training images to the test image. The keypoint transfer algorithm includes three steps: (i) keypoint matching, (ii) voting-based keypoint labeling, and (iii) keypoint-based probabilistic transfer of organ segmentations. We report segmentation results for abdominal organs in whole-body CT and MRI, as well as in contrast-enhanced CT and MRI. Our method offers a speed-up of about three orders of magnitude in comparison to common multi-atlas segmentation, while achieving an accuracy that compares favorably. Moreover, keypoint transfer does not require the registration to an atlas or a training phase. Finally, the method allows for the segmentation of scans with highly variable field-of-view.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge