Jonathan Rubin
LinguaMap: Which Layers of LLMs Speak Your Language and How to Tune Them?
Jan 27, 2026Abstract:Despite multilingual pretraining, large language models often struggle with non-English tasks, particularly in language control, the ability to respond in the intended language. We identify and characterize two key failure modes: the multilingual transfer bottleneck (correct language, incorrect task response) and the language consistency bottleneck (correct task response, wrong language). To systematically surface these issues, we design a four-scenario evaluation protocol spanning MMLU, MGSM, and XQuAD benchmarks. To probe these issues with interpretability, we extend logit lens analysis to track language probabilities layer by layer and compute cross-lingual semantic similarity of hidden states. The results reveal a three-phase internal structure: early layers align inputs into a shared semantic space, middle layers perform task reasoning, and late layers drive language-specific generation. Guided by these insights, we introduce selective fine-tuning of only the final layers responsible for language control. On Qwen-3-32B and Bloom-7.1B, this method achieves over 98 percent language consistency across six languages while fine-tuning only 3-5 percent of parameters, without sacrificing task accuracy. Importantly, this result is nearly identical to that of full-scope fine-tuning (for example, above 98 percent language consistency for both methods across all prompt scenarios) but uses a fraction of the computational resources. To the best of our knowledge, this is the first approach to leverage layer-localization of language control for efficient multilingual adaptation.
Limits of message passing for node classification: How class-bottlenecks restrict signal-to-noise ratio
Aug 25, 2025Abstract:Message passing neural networks (MPNNs) are powerful models for node classification but suffer from performance limitations under heterophily (low same-class connectivity) and structural bottlenecks in the graph. We provide a unifying statistical framework exposing the relationship between heterophily and bottlenecks through the signal-to-noise ratio (SNR) of MPNN representations. The SNR decomposes model performance into feature-dependent parameters and feature-independent sensitivities. We prove that the sensitivity to class-wise signals is bounded by higher-order homophily -- a generalisation of classical homophily to multi-hop neighbourhoods -- and show that low higher-order homophily manifests locally as the interaction between structural bottlenecks and class labels (class-bottlenecks). Through analysis of graph ensembles, we provide a further quantitative decomposition of bottlenecking into underreaching (lack of depth implying signals cannot arrive) and oversquashing (lack of breadth implying signals arriving on fewer paths) with closed-form expressions. We prove that optimal graph structures for maximising higher-order homophily are disjoint unions of single-class and two-class-bipartite clusters. This yields BRIDGE, a graph ensemble-based rewiring algorithm that achieves near-perfect classification accuracy across all homophily regimes on synthetic benchmarks and significant improvements on real-world benchmarks, by eliminating the ``mid-homophily pitfall'' where MPNNs typically struggle, surpassing current standard rewiring techniques from the literature. Our framework, whose code we make available for public use, provides both diagnostic tools for assessing MPNN performance, and simple yet effective methods for enhancing performance through principled graph modification.
Contrastive Self-Supervised Learning for Spatio-Temporal Analysis of Lung Ultrasound Videos
Oct 14, 2023Abstract:Self-supervised learning (SSL) methods have shown promise for medical imaging applications by learning meaningful visual representations, even when the amount of labeled data is limited. Here, we extend state-of-the-art contrastive learning SSL methods to 2D+time medical ultrasound video data by introducing a modified encoder and augmentation method capable of learning meaningful spatio-temporal representations, without requiring constraints on the input data. We evaluate our method on the challenging clinical task of identifying lung consolidations (an important pathological feature) in ultrasound videos. Using a multi-center dataset of over 27k lung ultrasound videos acquired from over 500 patients, we show that our method can significantly improve performance on downstream localization and classification of lung consolidation. Comparisons against baseline models trained without SSL show that the proposed methods are particularly advantageous when the size of labeled training data is limited (e.g., as little as 5% of the training set).
Convolution-Free Waveform Transformers for Multi-Lead ECG Classification
Sep 29, 2021



Abstract:We present our entry to the 2021 PhysioNet/CinC challenge - a waveform transformer model to detect cardiac abnormalities from ECG recordings. We compare the performance of the waveform transformer model on different ECG-lead subsets using approximately 88,000 ECG recordings from six datasets. In the official rankings, team prna ranked between 9 and 15 on 12, 6, 4, 3 and 2-lead sets respectively. Our waveform transformer model achieved an average challenge metric of 0.47 on the held-out test set across all ECG-lead subsets. Our combined performance across all leads placed us at rank 11 out of 39 officially ranking teams.
Interpretable Additive Recurrent Neural Networks For Multivariate Clinical Time Series
Sep 15, 2021Abstract:Time series models with recurrent neural networks (RNNs) can have high accuracy but are unfortunately difficult to interpret as a result of feature-interactions, temporal-interactions, and non-linear transformations. Interpretability is important in domains like healthcare where constructing models that provide insight into the relationships they have learned are required to validate and trust model predictions. We want accurate time series models where users can understand the contribution of individual input features. We present the Interpretable-RNN (I-RNN) that balances model complexity and accuracy by forcing the relationship between variables in the model to be additive. Interactions are restricted between hidden states of the RNN and additively combined at the final step. I-RNN specifically captures the unique characteristics of clinical time series, which are unevenly sampled in time, asynchronously acquired, and have missing data. Importantly, the hidden state activations represent feature coefficients that correlate with the prediction target and can be visualized as risk curves that capture the global relationship between individual input features and the outcome. We evaluate the I-RNN model on the Physionet 2012 Challenge dataset to predict in-hospital mortality, and on a real-world clinical decision support task: predicting hemodynamic interventions in the intensive care unit. I-RNN provides explanations in the form of global and local feature importances comparable to highly intelligible models like decision trees trained on hand-engineered features while significantly outperforming them. I-RNN remains intelligible while providing accuracy comparable to state-of-the-art decay-based and interpolation-based recurrent time series models. The experimental results on real-world clinical datasets refute the myth that there is a tradeoff between accuracy and interpretability.
CT-To-MR Conditional Generative Adversarial Networks for Ischemic Stroke Lesion Segmentation
Apr 30, 2019



Abstract:Infarcted brain tissue resulting from acute stroke readily shows up as hyperintense regions within diffusion-weighted magnetic resonance imaging (DWI). It has also been proposed that computed tomography perfusion (CTP) could alternatively be used to triage stroke patients, given improvements in speed and availability, as well as reduced cost. However, CTP has a lower signal to noise ratio compared to MR. In this work, we investigate whether a conditional mapping can be learned by a generative adversarial network to map CTP inputs to generated MR DWI that more clearly delineates hyperintense regions due to ischemic stroke. We detail the architectures of the generator and discriminator and describe the training process used to perform image-to-image translation from multi-modal CT perfusion maps to diffusion weighted MR outputs. We evaluate the results both qualitatively by visual comparison of generated MR to ground truth, as well as quantitatively by training fully convolutional neural networks that make use of generated MR data inputs to perform ischemic stroke lesion segmentation. Segmentation networks trained using generated CT-to-MR inputs result in at least some improvement on all metrics used for evaluation, compared with networks that only use CT perfusion input.
Semi-supervised Learning for Quantification of Pulmonary Edema in Chest X-Ray Images
Apr 10, 2019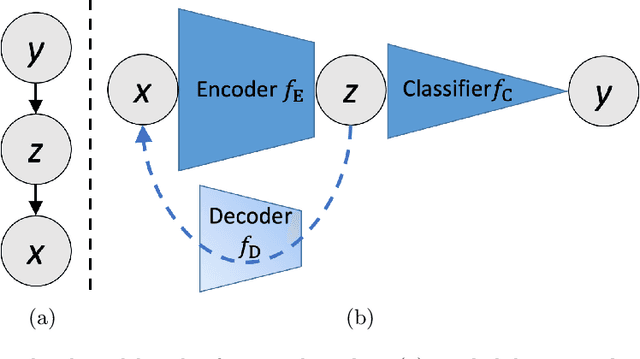
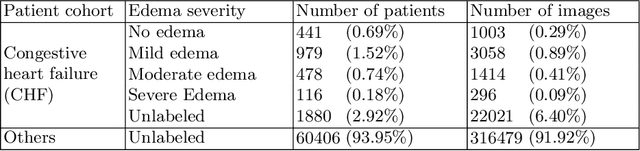
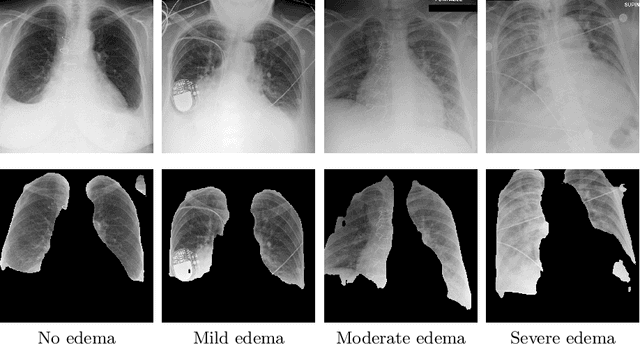
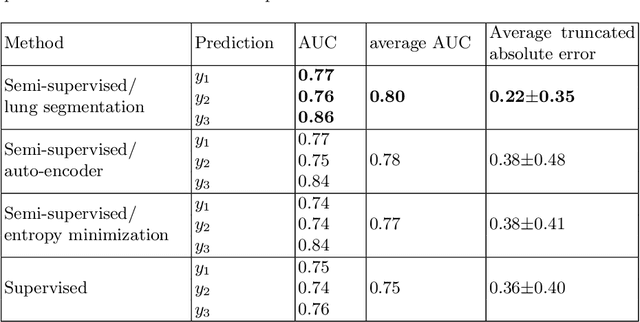
Abstract:We propose and demonstrate machine learning algorithms to assess the severity of pulmonary edema in chest x-ray images of congestive heart failure patients. Accurate assessment of pulmonary edema in heart failure is critical when making treatment and disposition decisions. Our work is grounded in a large-scale clinical dataset of over 300,000 x-ray images with associated radiology reports. While edema severity labels can be extracted unambiguously from a small fraction of the radiology reports, accurate annotation is challenging in most cases. To take advantage of the unlabeled images, we develop a Bayesian model that includes a variational auto-encoder for learning a latent representation from the entire image set trained jointly with a regressor that employs this representation for predicting pulmonary edema severity. Our experimental results suggest that modeling the distribution of images jointly with the limited labels improves the accuracy of pulmonary edema scoring compared to a strictly supervised approach. To the best of our knowledge, this is the first attempt to employ machine learning algorithms to automatically and quantitatively assess the severity of pulmonary edema in chest x-ray images.
Multivariate Time-series Similarity Assessment via Unsupervised Representation Learning and Stratified Locality Sensitive Hashing: Application to Early Acute Hypotensive Episode Detection
Dec 04, 2018



Abstract:Timely prediction of clinically critical events in Intensive Care Unit (ICU) is important for improving care and survival rate. Most of the existing approaches are based on the application of various classification methods on explicitly extracted statistical features from vital signals. In this work, we propose to eliminate the high cost of engineering hand-crafted features from multivariate time-series of physiologic signals by learning their representation with a sequence-to-sequence auto-encoder. We then propose to hash the learned representations to enable signal similarity assessment for the prediction of critical events. We apply this methodological framework to predict Acute Hypotensive Episodes (AHE) on a large and diverse dataset of vital signal recordings. Experiments demonstrate the ability of the presented framework in accurately predicting an upcoming AHE.
Ischemic Stroke Lesion Segmentation in CT Perfusion Scans using Pyramid Pooling and Focal Loss
Nov 02, 2018



Abstract:We present a fully convolutional neural network for segmenting ischemic stroke lesions in CT perfusion images for the ISLES 2018 challenge. Treatment of stroke is time sensitive and current standards for lesion identification require manual segmentation, a time consuming and challenging process. Automatic segmentation methods present the possibility of accurately identifying lesions and improving treatment planning. Our model is based on the PSPNet, a network architecture that makes use of pyramid pooling to provide global and local contextual information. To learn the varying shapes of the lesions, we train our network using focal loss, a loss function designed for the network to focus on learning the more difficult samples. We compare our model to networks trained using the U-Net and V-Net architectures. Our approach demonstrates effective performance in lesion segmentation and ranked among the top performers at the challenge conclusion.
Large Scale Automated Reading of Frontal and Lateral Chest X-Rays using Dual Convolutional Neural Networks
Apr 24, 2018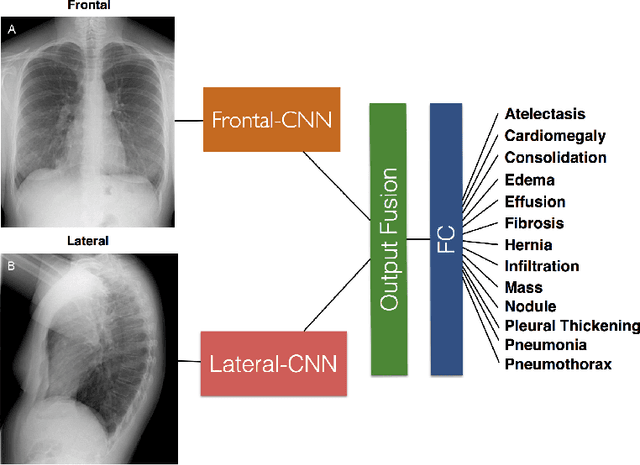
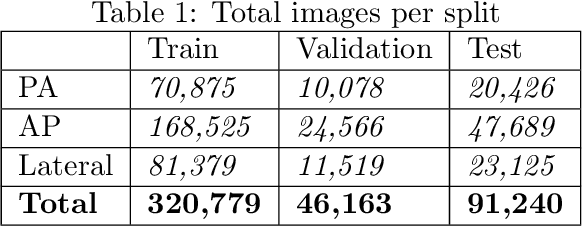
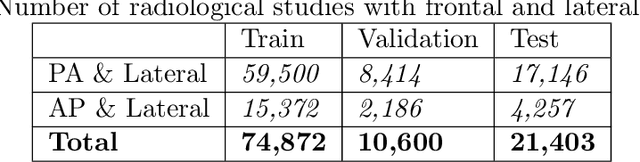
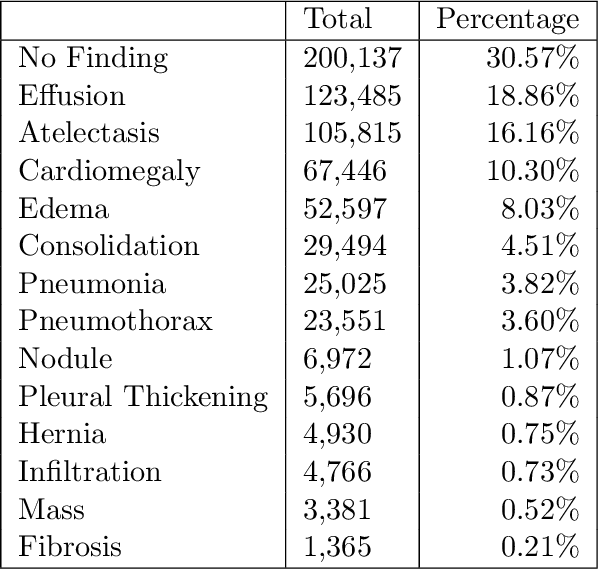
Abstract:The MIMIC-CXR dataset is (to date) the largest released chest x-ray dataset consisting of 473,064 chest x-rays and 206,574 radiology reports collected from 63,478 patients. We present the results of training and evaluating a collection of deep convolutional neural networks on this dataset to recognize multiple common thorax diseases. To the best of our knowledge, this is the first work that trains CNNs for this task on such a large collection of chest x-ray images, which is over four times the size of the largest previously released chest x-ray corpus (ChestX-Ray14). We describe and evaluate individual CNN models trained on frontal and lateral CXR view types. In addition, we present a novel DualNet architecture that emulates routine clinical practice by simultaneously processing both frontal and lateral CXR images obtained from a radiological exam. Our DualNet architecture shows improved performance in recognizing findings in CXR images when compared to applying separate baseline frontal and lateral classifiers.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge