Sourabh Kulhare
Contrastive Self-Supervised Learning for Spatio-Temporal Analysis of Lung Ultrasound Videos
Oct 14, 2023Abstract:Self-supervised learning (SSL) methods have shown promise for medical imaging applications by learning meaningful visual representations, even when the amount of labeled data is limited. Here, we extend state-of-the-art contrastive learning SSL methods to 2D+time medical ultrasound video data by introducing a modified encoder and augmentation method capable of learning meaningful spatio-temporal representations, without requiring constraints on the input data. We evaluate our method on the challenging clinical task of identifying lung consolidations (an important pathological feature) in ultrasound videos. Using a multi-center dataset of over 27k lung ultrasound videos acquired from over 500 patients, we show that our method can significantly improve performance on downstream localization and classification of lung consolidation. Comparisons against baseline models trained without SSL show that the proposed methods are particularly advantageous when the size of labeled training data is limited (e.g., as little as 5% of the training set).
How Good Are Synthetic Medical Images? An Empirical Study with Lung Ultrasound
Oct 05, 2023Abstract:Acquiring large quantities of data and annotations is known to be effective for developing high-performing deep learning models, but is difficult and expensive to do in the healthcare context. Adding synthetic training data using generative models offers a low-cost method to deal effectively with the data scarcity challenge, and can also address data imbalance and patient privacy issues. In this study, we propose a comprehensive framework that fits seamlessly into model development workflows for medical image analysis. We demonstrate, with datasets of varying size, (i) the benefits of generative models as a data augmentation method; (ii) how adversarial methods can protect patient privacy via data substitution; (iii) novel performance metrics for these use cases by testing models on real holdout data. We show that training with both synthetic and real data outperforms training with real data alone, and that models trained solely with synthetic data approach their real-only counterparts. Code is available at https://github.com/Global-Health-Labs/US-DCGAN.
Weakly Semi-Supervised Detection in Lung Ultrasound Videos
Aug 08, 2023Abstract:Frame-by-frame annotation of bounding boxes by clinical experts is often required to train fully supervised object detection models on medical video data. We propose a method for improving object detection in medical videos through weak supervision from video-level labels. More concretely, we aggregate individual detection predictions into video-level predictions and extend a teacher-student training strategy to provide additional supervision via a video-level loss. We also introduce improvements to the underlying teacher-student framework, including methods to improve the quality of pseudo-labels based on weak supervision and adaptive schemes to optimize knowledge transfer between the student and teacher networks. We apply this approach to the clinically important task of detecting lung consolidations (seen in respiratory infections such as COVID-19 pneumonia) in medical ultrasound videos. Experiments reveal that our framework improves detection accuracy and robustness compared to baseline semi-supervised models, and improves efficiency in data and annotation usage.
Fully-automated patient-level malaria assessment on field-prepared thin blood film microscopy images, including Supplementary Information
Aug 05, 2019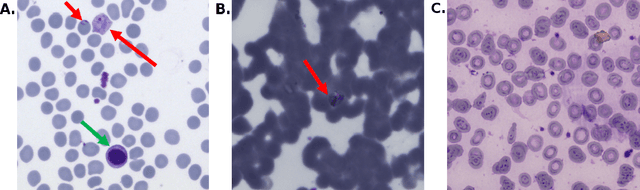
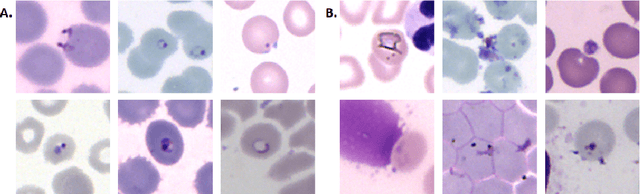
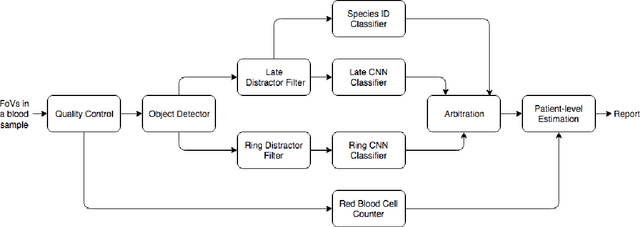
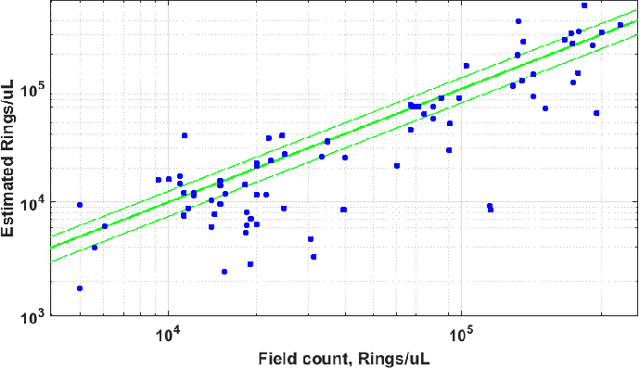
Abstract:Malaria is a life-threatening disease affecting millions. Microscopy-based assessment of thin blood films is a standard method to (i) determine malaria species and (ii) quantitate high-parasitemia infections. Full automation of malaria microscopy by machine learning (ML) is a challenging task because field-prepared slides vary widely in quality and presentation, and artifacts often heavily outnumber relatively rare parasites. In this work, we describe a complete, fully-automated framework for thin film malaria analysis that applies ML methods, including convolutional neural nets (CNNs), trained on a large and diverse dataset of field-prepared thin blood films. Quantitation and species identification results are close to sufficiently accurate for the concrete needs of drug resistance monitoring and clinical use-cases on field-prepared samples. We focus our methods and our performance metrics on the field use-case requirements. We discuss key issues and important metrics for the application of ML methods to malaria microscopy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge