Mayoore S. Jaiswal
Large Scale Neural Architecture Search with Polyharmonic Splines
Nov 20, 2020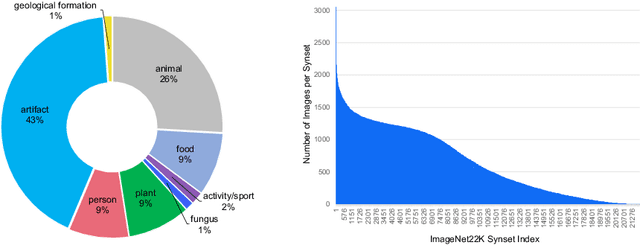
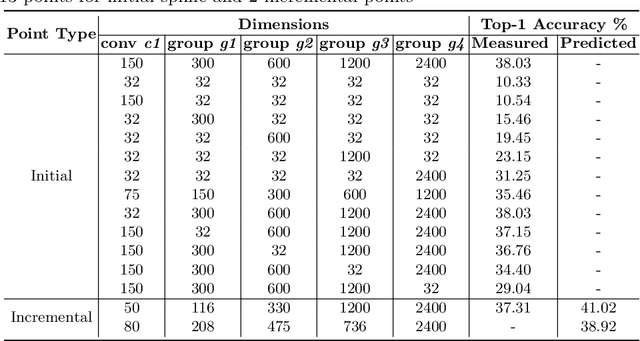
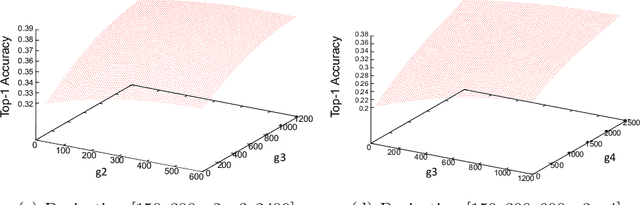
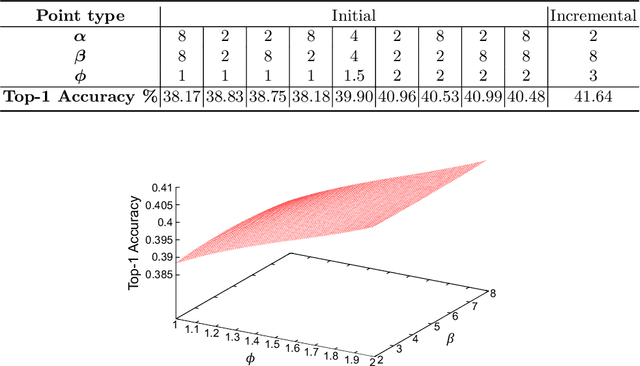
Abstract:Neural Architecture Search (NAS) is a powerful tool to automatically design deep neural networks for many tasks, including image classification. Due to the significant computational burden of the search phase, most NAS methods have focused so far on small, balanced datasets. All attempts at conducting NAS at large scale have employed small proxy sets, and then transferred the learned architectures to larger datasets by replicating or stacking the searched cells. We propose a NAS method based on polyharmonic splines that can perform search directly on large scale, imbalanced target datasets. We demonstrate the effectiveness of our method on the ImageNet22K benchmark[16], which contains 14 million images distributed in a highly imbalanced manner over 21,841 categories. By exploring the search space of the ResNet [23] and Big-Little Net ResNext [11] architectures directly on ImageNet22K, our polyharmonic splines NAS method designed a model which achieved a top-1 accuracy of 40.03% on ImageNet22K, an absolute improvement of 3.13% over the state of the art with similar global batch size [15].
Ideas for Improving the Field of Machine Learning: Summarizing Discussion from the NeurIPS 2019 Retrospectives Workshop
Jul 21, 2020Abstract:This report documents ideas for improving the field of machine learning, which arose from discussions at the ML Retrospectives workshop at NeurIPS 2019. The goal of the report is to disseminate these ideas more broadly, and in turn encourage continuing discussion about how the field could improve along these axes. We focus on topics that were most discussed at the workshop: incentives for encouraging alternate forms of scholarship, re-structuring the review process, participation from academia and industry, and how we might better train computer scientists as scientists. Videos from the workshop can be accessed at https://slideslive.com/neurips/west-114-115-retrospectives-a-venue-for-selfreflection-in-ml-research
MUTE: Data-Similarity Driven Multi-hot Target Encoding for Neural Network Design
Oct 15, 2019



Abstract:Target encoding is an effective technique to deliver better performance for conventional machine learning methods, and recently, for deep neural networks as well. However, the existing target encoding approaches require significant increase in the learning capacity, thus demand higher computation power and more training data. In this paper, we present a novel and efficient target encoding scheme, MUTE to improve both generalizability and robustness of a target model by understanding the inter-class characteristics of a target dataset. By extracting the confusion level between the target classes in a dataset, MUTE strategically optimizes the Hamming distances among target encoding. Such optimized target encoding offers higher classification strength for neural network models with negligible computation overhead and without increasing the model size. When MUTE is applied to the popular image classification networks and datasets, our experimental results show that MUTE offers better generalization and defense against the noises and adversarial attacks over the existing solutions.
Fully-automated patient-level malaria assessment on field-prepared thin blood film microscopy images, including Supplementary Information
Aug 05, 2019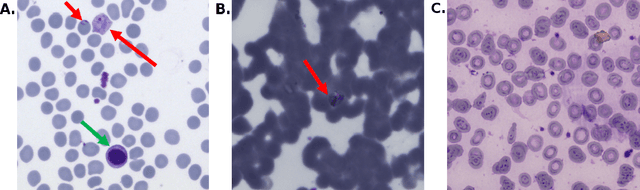
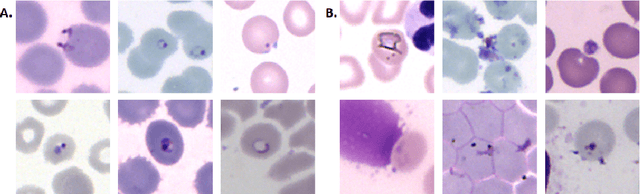
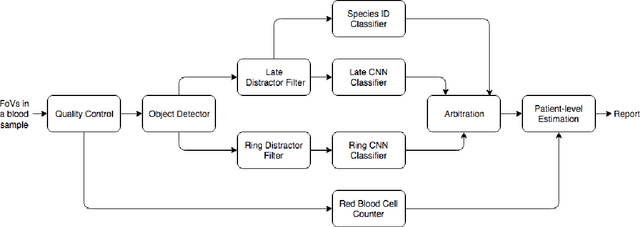
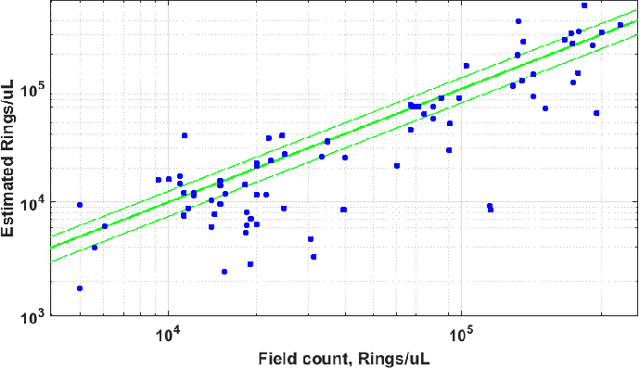
Abstract:Malaria is a life-threatening disease affecting millions. Microscopy-based assessment of thin blood films is a standard method to (i) determine malaria species and (ii) quantitate high-parasitemia infections. Full automation of malaria microscopy by machine learning (ML) is a challenging task because field-prepared slides vary widely in quality and presentation, and artifacts often heavily outnumber relatively rare parasites. In this work, we describe a complete, fully-automated framework for thin film malaria analysis that applies ML methods, including convolutional neural nets (CNNs), trained on a large and diverse dataset of field-prepared thin blood films. Quantitation and species identification results are close to sufficiently accurate for the concrete needs of drug resistance monitoring and clinical use-cases on field-prepared samples. We focus our methods and our performance metrics on the field use-case requirements. We discuss key issues and important metrics for the application of ML methods to malaria microscopy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge