Matthew P. Horning
Beyond validation loss: Clinically-tailored optimization metrics improve a model's clinical performance
Jan 22, 2026Abstract:A key task in ML is to optimize models at various stages, e.g. by choosing hyperparameters or picking a stopping point. A traditional ML approach is to use validation loss, i.e. to apply the training loss function on a validation set to guide these optimizations. However, ML for healthcare has a distinct goal from traditional ML: Models must perform well relative to specific clinical requirements, vs. relative to the loss function used for training. These clinical requirements can be captured more precisely by tailored metrics. Since many optimization tasks do not require the driving metric to be differentiable, they allow a wider range of options, including the use of metrics tailored to be clinically-relevant. In this paper we describe two controlled experiments which show how the use of clinically-tailored metrics provide superior model optimization compared to validation loss, in the sense of better performance on the clinical task. The use of clinically-relevant metrics for optimization entails some extra effort, to define the metrics and to code them into the pipeline. But it can yield models that better meet the central goal of ML for healthcare: strong performance in the clinic.
Driving down Poisson error can offset classification error in clinical tasks
May 09, 2024Abstract:Medical machine learning algorithms are typically evaluated based on accuracy vs. a clinician-defined ground truth, a reasonable choice because trained clinicians are usually better classifiers than ML models. However, this metric does not fully reflect the clinical task: it neglects the fact that humans, even with perfect accuracy, are subject to sometimes significant error from the Poisson statistics of rare events, because clinical protocols often specify that a relatively small sample be examined. For example, to quantitate malaria on a thin blood film a clinician examines only 2000 red blood cells (0.0004 uL), which can yield large variation in actual number of parasites present due to Poisson variability, so that a perfect human's count can differ substantially from the true average load. In contrast, ML systems may be less accurate on an object level, but they also may have the option to examine more blood (e.g. 0.1 uL, or 250x). So while their accuracy as to parasite count in a particular sample is lower, the Poisson variability of their estimate is also lower due to larger sample size. Crucially, when an ML system moves out of the proof-of-concept stage and targets deployment in a clinical setting, its performance must match current standard of care. To this end, it may have the option to offset its lower accuracy by increasing sample size to reduce Poisson error, and thus attain the same net clinical performance as a perfectly accurate human limited by smaller sample size. In this paper, we analyze the mathematics of the trade-off between these two types of error, to enable teams developing ML systems to leverage a relative strength (larger sample sizes) to offset a relative weakness (classification accuracy). We illustrate the methods with two concrete examples: diagnosis and quantitation of malaria on blood films.
Use case-focused metrics to evaluate machine learning for diseases involving parasite loads
Sep 14, 2022


Abstract:Communal hill-climbing, via comparison of algorithm performances, can greatly accelerate ML research. However, it requires task-relevant metrics. For diseases involving parasite loads, e.g., malaria and neglected tropical diseases (NTDs) such as schistosomiasis, the metrics currently reported in ML papers (e.g., AUC, F1 score) are ill-suited to the clinical task. As a result, the hill-climbing system is not enabling progress towards solutions that address these dire illnesses. Drawing on examples from malaria and NTDs, this paper highlights two gaps in current ML practice and proposes methods for improvement: (i) We describe aspects of ML development, and performance metrics in particular, that need to be firmly grounded in the clinical use case, and we offer methods for acquiring this domain knowledge. (ii) We describe in detail performance metrics to guide development of ML models for diseases involving parasite loads. We highlight the importance of a patient-level perspective, interpatient variability, false positive rates, limit of detection, and different types of error. We also discuss problems with ROC curves and AUC as commonly used in this context.
Fully-automated patient-level malaria assessment on field-prepared thin blood film microscopy images, including Supplementary Information
Aug 05, 2019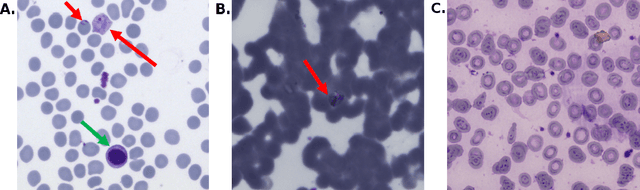
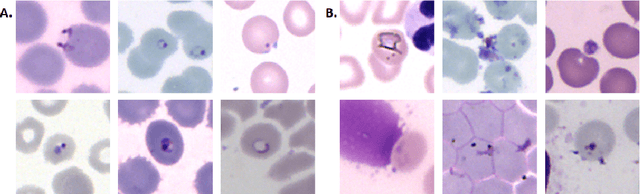
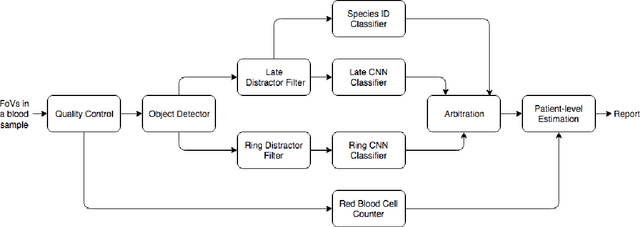
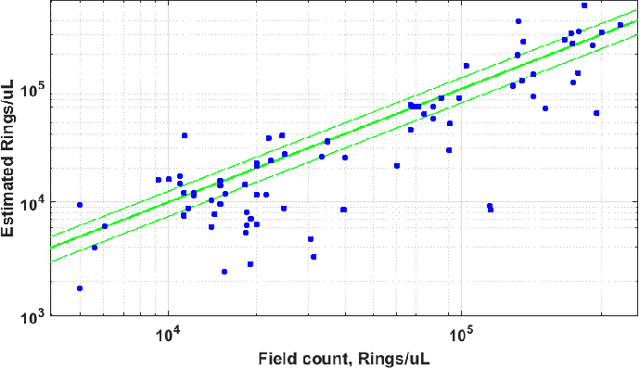
Abstract:Malaria is a life-threatening disease affecting millions. Microscopy-based assessment of thin blood films is a standard method to (i) determine malaria species and (ii) quantitate high-parasitemia infections. Full automation of malaria microscopy by machine learning (ML) is a challenging task because field-prepared slides vary widely in quality and presentation, and artifacts often heavily outnumber relatively rare parasites. In this work, we describe a complete, fully-automated framework for thin film malaria analysis that applies ML methods, including convolutional neural nets (CNNs), trained on a large and diverse dataset of field-prepared thin blood films. Quantitation and species identification results are close to sufficiently accurate for the concrete needs of drug resistance monitoring and clinical use-cases on field-prepared samples. We focus our methods and our performance metrics on the field use-case requirements. We discuss key issues and important metrics for the application of ML methods to malaria microscopy.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge