Sarah Frisken
Rapid patient-specific neural networks for intraoperative X-ray to volume registration
Mar 20, 2025Abstract:The integration of artificial intelligence in image-guided interventions holds transformative potential, promising to extract 3D geometric and quantitative information from conventional 2D imaging modalities during complex procedures. Achieving this requires the rapid and precise alignment of 2D intraoperative images (e.g., X-ray) with 3D preoperative volumes (e.g., CT, MRI). However, current 2D/3D registration methods fail across the broad spectrum of procedures dependent on X-ray guidance: traditional optimization techniques require custom parameter tuning for each subject, whereas neural networks trained on small datasets do not generalize to new patients or require labor-intensive manual annotations, increasing clinical burden and precluding application to new anatomical targets. To address these challenges, we present xvr, a fully automated framework for training patient-specific neural networks for 2D/3D registration. xvr uses physics-based simulation to generate abundant high-quality training data from a patient's own preoperative volumetric imaging, thereby overcoming the inherently limited ability of supervised models to generalize to new patients and procedures. Furthermore, xvr requires only 5 minutes of training per patient, making it suitable for emergency interventions as well as planned procedures. We perform the largest evaluation of a 2D/3D registration algorithm on real X-ray data to date and find that xvr robustly generalizes across a diverse dataset comprising multiple anatomical structures, imaging modalities, and hospitals. Across surgical tasks, xvr achieves submillimeter-accurate registration at intraoperative speeds, improving upon existing methods by an order of magnitude. xvr is released as open-source software freely available at https://github.com/eigenvivek/xvr.
3D/2D Registration of Angiograms using Silhouette-based Differentiable Rendering
Jan 24, 2025Abstract:We present a method for 3D/2D registration of Digital Subtraction Angiography (DSA) images to provide valuable insight into brain hemodynamics and angioarchitecture. Our approach formulates the registration as a pose estimation problem, leveraging both anteroposterior and lateral DSA views and employing differentiable rendering. Preliminary experiments on real and synthetic datasets demonstrate the effectiveness of our method, with both qualitative and quantitative evaluations highlighting its potential for clinical applications. The code is available at https://github.com/taewoonglee17/TwoViewsDSAReg.
Differentiable Voxel-based X-ray Rendering Improves Sparse-View 3D CBCT Reconstruction
Dec 02, 2024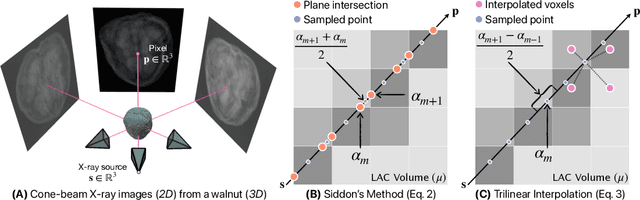
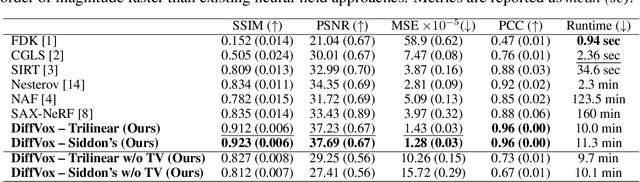
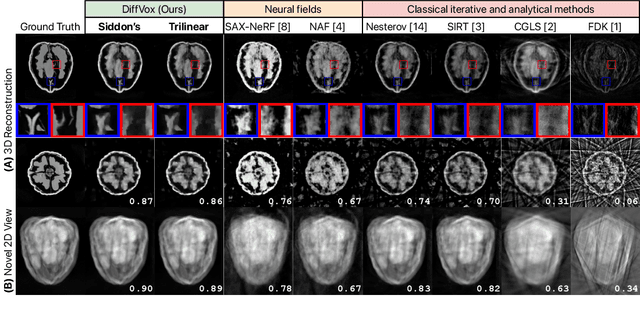
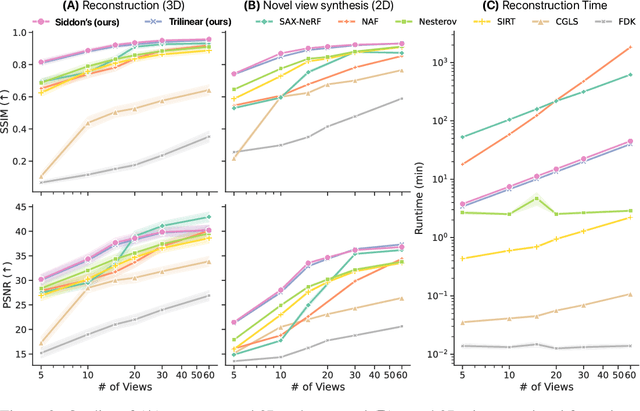
Abstract:We present DiffVox, a self-supervised framework for Cone-Beam Computed Tomography (CBCT) reconstruction by directly optimizing a voxelgrid representation using physics-based differentiable X-ray rendering. Further, we investigate how the different implementations of the X-ray image formation model in the renderer affect the quality of 3D reconstruction and novel view synthesis. When combined with our regularized voxel-based learning framework, we find that using an exact implementation of the discrete Beer-Lambert law for X-ray attenuation in the renderer outperforms both widely used iterative CBCT reconstruction algorithms and modern neural field approaches, particularly when given only a few input views. As a result, we reconstruct high-fidelity 3D CBCT volumes from fewer X-rays, potentially reducing ionizing radiation exposure and improving diagnostic utility. Our implementation is available at https://github.com/hossein-momeni/DiffVox.
Unified Cross-Modal Image Synthesis with Hierarchical Mixture of Product-of-Experts
Oct 25, 2024Abstract:We propose a deep mixture of multimodal hierarchical variational auto-encoders called MMHVAE that synthesizes missing images from observed images in different modalities. MMHVAE's design focuses on tackling four challenges: (i) creating a complex latent representation of multimodal data to generate high-resolution images; (ii) encouraging the variational distributions to estimate the missing information needed for cross-modal image synthesis; (iii) learning to fuse multimodal information in the context of missing data; (iv) leveraging dataset-level information to handle incomplete data sets at training time. Extensive experiments are performed on the challenging problem of pre-operative brain multi-parametric magnetic resonance and intra-operative ultrasound imaging.
Intraoperative Registration by Cross-Modal Inverse Neural Rendering
Sep 18, 2024Abstract:We present in this paper a novel approach for 3D/2D intraoperative registration during neurosurgery via cross-modal inverse neural rendering. Our approach separates implicit neural representation into two components, handling anatomical structure preoperatively and appearance intraoperatively. This disentanglement is achieved by controlling a Neural Radiance Field's appearance with a multi-style hypernetwork. Once trained, the implicit neural representation serves as a differentiable rendering engine, which can be used to estimate the surgical camera pose by minimizing the dissimilarity between its rendered images and the target intraoperative image. We tested our method on retrospective patients' data from clinical cases, showing that our method outperforms state-of-the-art while meeting current clinical standards for registration. Code and additional resources can be found at https://maxfehrentz.github.io/style-ngp/.
Learning to Match 2D Keypoints Across Preoperative MR and Intraoperative Ultrasound
Sep 12, 2024Abstract:We propose in this paper a texture-invariant 2D keypoints descriptor specifically designed for matching preoperative Magnetic Resonance (MR) images with intraoperative Ultrasound (US) images. We introduce a matching-by-synthesis strategy, where intraoperative US images are synthesized from MR images accounting for multiple MR modalities and intraoperative US variability. We build our training set by enforcing keypoints localization over all images then train a patient-specific descriptor network that learns texture-invariant discriminant features in a supervised contrastive manner, leading to robust keypoints descriptors. Our experiments on real cases with ground truth show the effectiveness of the proposed approach, outperforming the state-of-the-art methods and achieving 80.35% matching precision on average.
Patient-Specific Real-Time Segmentation in Trackerless Brain Ultrasound
May 16, 2024



Abstract:Intraoperative ultrasound (iUS) imaging has the potential to improve surgical outcomes in brain surgery. However, its interpretation is challenging, even for expert neurosurgeons. In this work, we designed the first patient-specific framework that performs brain tumor segmentation in trackerless iUS. To disambiguate ultrasound imaging and adapt to the neurosurgeon's surgical objective, a patient-specific real-time network is trained using synthetic ultrasound data generated by simulating virtual iUS sweep acquisitions in pre-operative MR data. Extensive experiments performed in real ultrasound data demonstrate the effectiveness of the proposed approach, allowing for adapting to the surgeon's definition of surgical targets and outperforming non-patient-specific models, neurosurgeon experts, and high-end tracking systems. Our code is available at: \url{https://github.com/ReubenDo/MHVAE-Seg}.
Spatiotemporal Disentanglement of Arteriovenous Malformations in Digital Subtraction Angiography
Feb 15, 2024Abstract:Although Digital Subtraction Angiography (DSA) is the most important imaging for visualizing cerebrovascular anatomy, its interpretation by clinicians remains difficult. This is particularly true when treating arteriovenous malformations (AVMs), where entangled vasculature connecting arteries and veins needs to be carefully identified.The presented method aims to enhance DSA image series by highlighting critical information via automatic classification of vessels using a combination of two learning models: An unsupervised machine learning method based on Independent Component Analysis that decomposes the phases of flow and a convolutional neural network that automatically delineates the vessels in image space. The proposed method was tested on clinical DSA images series and demonstrated efficient differentiation between arteries and veins that provides a viable solution to enhance visualizations for clinical use.
Learning Expected Appearances for Intraoperative Registration during Neurosurgery
Oct 03, 2023Abstract:We present a novel method for intraoperative patient-to-image registration by learning Expected Appearances. Our method uses preoperative imaging to synthesize patient-specific expected views through a surgical microscope for a predicted range of transformations. Our method estimates the camera pose by minimizing the dissimilarity between the intraoperative 2D view through the optical microscope and the synthesized expected texture. In contrast to conventional methods, our approach transfers the processing tasks to the preoperative stage, reducing thereby the impact of low-resolution, distorted, and noisy intraoperative images, that often degrade the registration accuracy. We applied our method in the context of neuronavigation during brain surgery. We evaluated our approach on synthetic data and on retrospective data from 6 clinical cases. Our method outperformed state-of-the-art methods and achieved accuracies that met current clinical standards.
Unified Brain MR-Ultrasound Synthesis using Multi-Modal Hierarchical Representations
Sep 19, 2023Abstract:We introduce MHVAE, a deep hierarchical variational auto-encoder (VAE) that synthesizes missing images from various modalities. Extending multi-modal VAEs with a hierarchical latent structure, we introduce a probabilistic formulation for fusing multi-modal images in a common latent representation while having the flexibility to handle incomplete image sets as input. Moreover, adversarial learning is employed to generate sharper images. Extensive experiments are performed on the challenging problem of joint intra-operative ultrasound (iUS) and Magnetic Resonance (MR) synthesis. Our model outperformed multi-modal VAEs, conditional GANs, and the current state-of-the-art unified method (ResViT) for synthesizing missing images, demonstrating the advantage of using a hierarchical latent representation and a principled probabilistic fusion operation. Our code is publicly available \url{https://github.com/ReubenDo/MHVAE}.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge