Alexandra Golby
Unified Cross-Modal Image Synthesis with Hierarchical Mixture of Product-of-Experts
Oct 25, 2024Abstract:We propose a deep mixture of multimodal hierarchical variational auto-encoders called MMHVAE that synthesizes missing images from observed images in different modalities. MMHVAE's design focuses on tackling four challenges: (i) creating a complex latent representation of multimodal data to generate high-resolution images; (ii) encouraging the variational distributions to estimate the missing information needed for cross-modal image synthesis; (iii) learning to fuse multimodal information in the context of missing data; (iv) leveraging dataset-level information to handle incomplete data sets at training time. Extensive experiments are performed on the challenging problem of pre-operative brain multi-parametric magnetic resonance and intra-operative ultrasound imaging.
Intraoperative Registration by Cross-Modal Inverse Neural Rendering
Sep 18, 2024Abstract:We present in this paper a novel approach for 3D/2D intraoperative registration during neurosurgery via cross-modal inverse neural rendering. Our approach separates implicit neural representation into two components, handling anatomical structure preoperatively and appearance intraoperatively. This disentanglement is achieved by controlling a Neural Radiance Field's appearance with a multi-style hypernetwork. Once trained, the implicit neural representation serves as a differentiable rendering engine, which can be used to estimate the surgical camera pose by minimizing the dissimilarity between its rendered images and the target intraoperative image. We tested our method on retrospective patients' data from clinical cases, showing that our method outperforms state-of-the-art while meeting current clinical standards for registration. Code and additional resources can be found at https://maxfehrentz.github.io/style-ngp/.
Learning to Match 2D Keypoints Across Preoperative MR and Intraoperative Ultrasound
Sep 12, 2024Abstract:We propose in this paper a texture-invariant 2D keypoints descriptor specifically designed for matching preoperative Magnetic Resonance (MR) images with intraoperative Ultrasound (US) images. We introduce a matching-by-synthesis strategy, where intraoperative US images are synthesized from MR images accounting for multiple MR modalities and intraoperative US variability. We build our training set by enforcing keypoints localization over all images then train a patient-specific descriptor network that learns texture-invariant discriminant features in a supervised contrastive manner, leading to robust keypoints descriptors. Our experiments on real cases with ground truth show the effectiveness of the proposed approach, outperforming the state-of-the-art methods and achieving 80.35% matching precision on average.
Patient-Specific Real-Time Segmentation in Trackerless Brain Ultrasound
May 16, 2024



Abstract:Intraoperative ultrasound (iUS) imaging has the potential to improve surgical outcomes in brain surgery. However, its interpretation is challenging, even for expert neurosurgeons. In this work, we designed the first patient-specific framework that performs brain tumor segmentation in trackerless iUS. To disambiguate ultrasound imaging and adapt to the neurosurgeon's surgical objective, a patient-specific real-time network is trained using synthetic ultrasound data generated by simulating virtual iUS sweep acquisitions in pre-operative MR data. Extensive experiments performed in real ultrasound data demonstrate the effectiveness of the proposed approach, allowing for adapting to the surgeon's definition of surgical targets and outperforming non-patient-specific models, neurosurgeon experts, and high-end tracking systems. Our code is available at: \url{https://github.com/ReubenDo/MHVAE-Seg}.
Spatiotemporal Disentanglement of Arteriovenous Malformations in Digital Subtraction Angiography
Feb 15, 2024Abstract:Although Digital Subtraction Angiography (DSA) is the most important imaging for visualizing cerebrovascular anatomy, its interpretation by clinicians remains difficult. This is particularly true when treating arteriovenous malformations (AVMs), where entangled vasculature connecting arteries and veins needs to be carefully identified.The presented method aims to enhance DSA image series by highlighting critical information via automatic classification of vessels using a combination of two learning models: An unsupervised machine learning method based on Independent Component Analysis that decomposes the phases of flow and a convolutional neural network that automatically delineates the vessels in image space. The proposed method was tested on clinical DSA images series and demonstrated efficient differentiation between arteries and veins that provides a viable solution to enhance visualizations for clinical use.
Unified Brain MR-Ultrasound Synthesis using Multi-Modal Hierarchical Representations
Sep 19, 2023Abstract:We introduce MHVAE, a deep hierarchical variational auto-encoder (VAE) that synthesizes missing images from various modalities. Extending multi-modal VAEs with a hierarchical latent structure, we introduce a probabilistic formulation for fusing multi-modal images in a common latent representation while having the flexibility to handle incomplete image sets as input. Moreover, adversarial learning is employed to generate sharper images. Extensive experiments are performed on the challenging problem of joint intra-operative ultrasound (iUS) and Magnetic Resonance (MR) synthesis. Our model outperformed multi-modal VAEs, conditional GANs, and the current state-of-the-art unified method (ResViT) for synthesizing missing images, demonstrating the advantage of using a hierarchical latent representation and a principled probabilistic fusion operation. Our code is publicly available \url{https://github.com/ReubenDo/MHVAE}.
Do Public Datasets Assure Unbiased Comparisons for Registration Evaluation?
Mar 20, 2020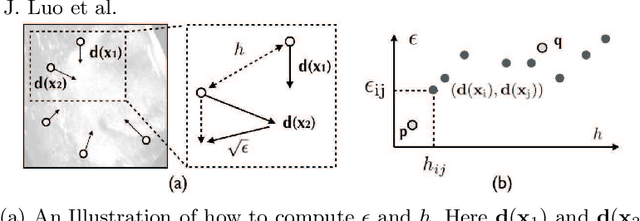
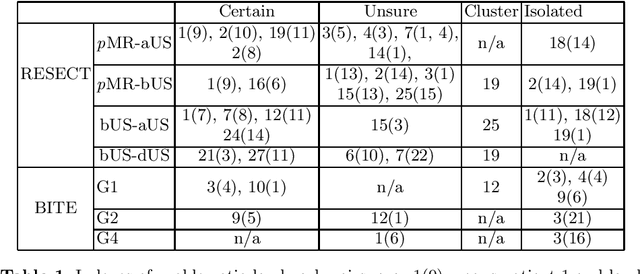
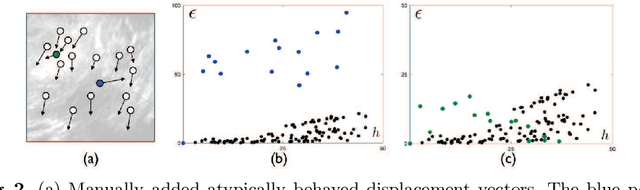
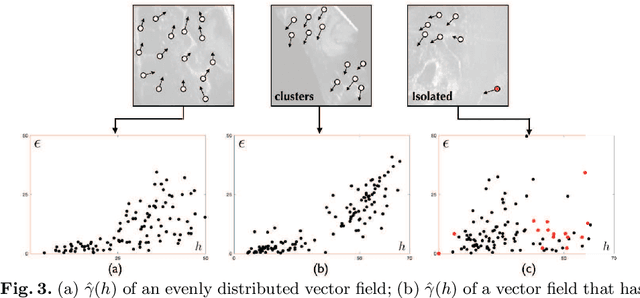
Abstract:With the increasing availability of new image registration approaches, an unbiased evaluation is becoming more needed so that clinicians can choose the most suitable approaches for their applications. Current evaluations typically use landmarks in manually annotated datasets. As a result, the quality of annotations is crucial for unbiased comparisons. Even though most data providers claim to have quality control over their datasets, an objective third-party screening can be reassuring for intended users. In this study, we use the variogram to screen the manually annotated landmarks in two datasets used to benchmark registration in image-guided neurosurgeries. The variogram provides an intuitive 2D representation of the spatial characteristics of annotated landmarks. Using variograms, we identified potentially problematic cases and had them examined by experienced radiologists. We found that (1) a small number of annotations may have fiducial localization errors; (2) the landmark distribution for some cases is not ideal to offer fair comparisons. If unresolved, both findings could incur bias in registration evaluation.
Pilot Study on Verifying the Monotonic Relationship between Error and Uncertainty in Deformable Registration for Neurosurgery
Aug 21, 2019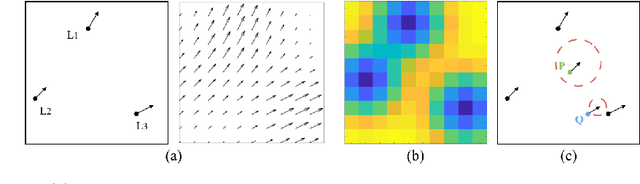
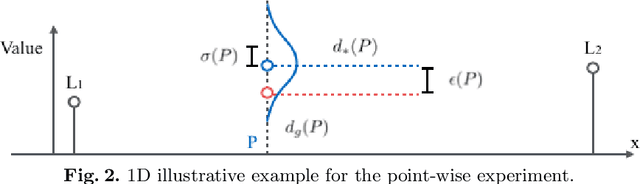
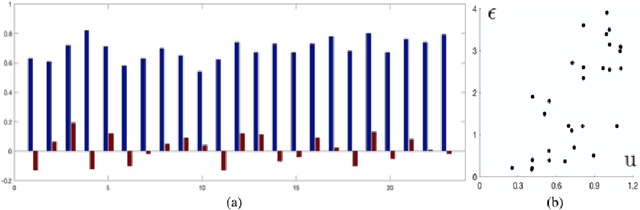
Abstract:In image-guided neurosurgery, deformable registration currently is not a clinical routine. Although using it in practice is a goal for image-guided therapy, this goal is hampered because surgeons are wary of the less predictable deformable registration error. In the preoperative- to-intraoperative registration, when surgeons notice a misaligned image pattern, they want to know whether it is a registration error or an actual deformation caused by tumor resection or retraction. Here, surgeons need a spatial distribution of error to help them make a better-informed decision, i.e., ignore locations with high error. However, such an error estimate is difficult to acquire. Alternatively, probabilistic image registration (PIR) methods give measures of registration uncertainty, which is a potential surrogate for assessing the quality of registration results. It is intuitive and believed by a lot of people that high uncertainty indicates a large error. Yet to the best of our knowledge, no such conclusion has been reported in the PIR literature. In this study, we look at one PIR method and give preliminary results showing that point-wise registration error and uncertainty are monotonically correlated.
On the Ambiguity of Registration Uncertainty
Mar 20, 2018



Abstract:Estimating the uncertainty in image registration is an area of current research that is aimed at providing information that will enable surgeons to assess the operative risk based on registered image data and the estimated registration uncertainty. If they receive inaccurately calculated registration uncertainty and misplace confidence in the alignment solutions, severe consequences may result. For probabilistic image registration (PIR), most research quantifies the registration uncertainty using summary statistics of the transformation distributions. In this paper, we study a rarely examined topic: whether those summary statistics of the transformation distribution truly represent the registration uncertainty. Using concrete examples, we show that there are two types of uncertainties: the transformation uncertainty, Ut, and label uncertainty Ul. Ut indicates the doubt concerning transformation parameters and can be estimated by conventional uncertainty measures, while Ul is strongly linked to the goal of registration. Further, we show that using Ut to quantify Ul is inappropriate and can be misleading. In addition, we present some potentially critical findings regarding PIR.
A Feature-Driven Active Framework for Ultrasound-Based Brain Shift Compensation
Mar 20, 2018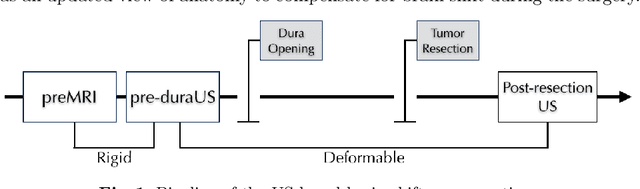
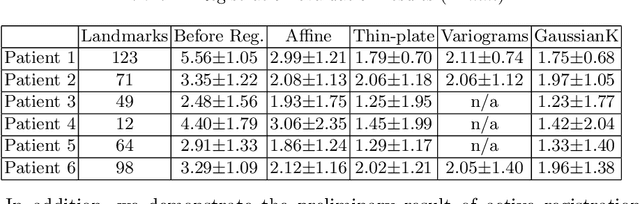
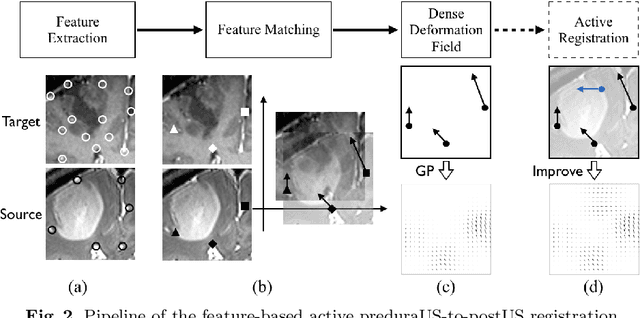
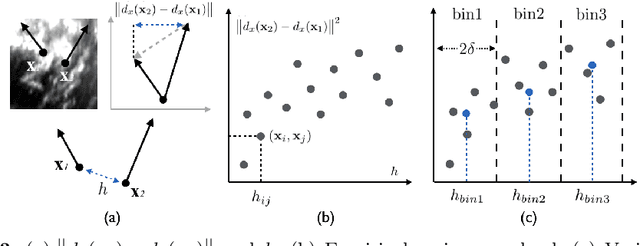
Abstract:A reliable Ultrasound (US)-to-US registration method to compensate for brain shift would substantially improve Image-Guided Neurological Surgery. Developing such a registration method is very challenging, due to factors such as missing correspondence in images, the complexity of brain pathology and the demand for fast computation. We propose a novel feature-driven active framework. Here, landmarks and their displacement are first estimated from a pair of US images using corresponding local image features. Subsequently, a Gaussian Process (GP) model is used to interpolate a dense deformation field from the sparse landmarks. Kernels of the GP are estimated by using variograms and a discrete grid search method. If necessary, the user can actively add new landmarks based on the image context and visualization of the uncertainty measure provided by the GP to further improve the result. We retrospectively demonstrate our registration framework as a robust and accurate brain shift compensation solution on clinical data acquired during neurosurgery.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge