Miaomiao Zhang
Anatomically Guided Latent Diffusion for Brain MRI Progression Modeling
Jan 21, 2026Abstract:Accurately modeling longitudinal brain MRI progression is crucial for understanding neurodegenerative diseases and predicting individualized structural changes. Existing state-of-the-art approaches, such as Brain Latent Progression (BrLP), often use multi-stage training pipelines with auxiliary conditioning modules but suffer from architectural complexity, suboptimal use of conditional clinical covariates, and limited guarantees of anatomical consistency. We propose Anatomically Guided Latent Diffusion Model (AG-LDM), a segmentation-guided framework that enforces anatomically consistent progression while substantially simplifying the training pipeline. AG-LDM conditions latent diffusion by directly fusing baseline anatomy, noisy follow-up states, and clinical covariates at the input level, a strategy that avoids auxiliary control networks by learning a unified, end-to-end model that represents both anatomy and progression. A lightweight 3D tissue segmentation model (WarpSeg) provides explicit anatomical supervision during both autoencoder fine-tuning and diffusion model training, ensuring consistent brain tissue boundaries and morphometric fidelity. Experiments on 31,713 ADNI longitudinal pairs and zero-shot evaluation on OASIS-3 demonstrate that AG-LDM matches or surpasses more complex diffusion models, achieving state-of-the-art image quality and 15-20\% reduction in volumetric errors in generated images. AG-LDM also exhibits markedly stronger utilization of temporal and clinical covariates (up to 31.5x higher sensitivity than BrLP) and generates biologically plausible counterfactual trajectories, accurately capturing hallmarks of Alzheimer's progression such as limbic atrophy and ventricular expansion. These results highlight AG-LDM as an efficient, anatomically grounded framework for reliable brain MRI progression modeling.
Who is a Better Matchmaker? Human vs. Algorithmic Judge Assignment in a High-Stakes Startup Competition
Oct 14, 2025



Abstract:There is growing interest in applying artificial intelligence (AI) to automate and support complex decision-making tasks. However, it remains unclear how algorithms compare to human judgment in contexts requiring semantic understanding and domain expertise. We examine this in the context of the judge assignment problem, matching submissions to suitably qualified judges. Specifically, we tackled this problem at the Harvard President's Innovation Challenge, the university's premier venture competition awarding over \$500,000 to student and alumni startups. This represents a real-world environment where high-quality judge assignment is essential. We developed an AI-based judge-assignment algorithm, Hybrid Lexical-Semantic Similarity Ensemble (HLSE), and deployed it at the competition. We then evaluated its performance against human expert assignments using blinded match-quality scores from judges on $309$ judge-venture pairs. Using a Mann-Whitney U statistic based test, we found no statistically significant difference in assignment quality between the two approaches ($AUC=0.48, p=0.40$); on average, algorithmic matches are rated $3.90$ and manual matches $3.94$ on a 5-point scale, where 5 indicates an excellent match. Furthermore, manual assignments that previously required a full week could be automated in several hours by the algorithm during deployment. These results demonstrate that HLSE achieves human-expert-level matching quality while offering greater scalability and efficiency, underscoring the potential of AI-driven solutions to support and enhance human decision-making for judge assignment in high-stakes settings.
Control Synthesis of Cyber-Physical Systems for Real-Time Specifications through Causation-Guided Reinforcement Learning
Oct 09, 2025Abstract:In real-time and safety-critical cyber-physical systems (CPSs), control synthesis must guarantee that generated policies meet stringent timing and correctness requirements under uncertain and dynamic conditions. Signal temporal logic (STL) has emerged as a powerful formalism of expressing real-time constraints, with its semantics enabling quantitative assessment of system behavior. Meanwhile, reinforcement learning (RL) has become an important method for solving control synthesis problems in unknown environments. Recent studies incorporate STL-based reward functions into RL to automatically synthesize control policies. However, the automatically inferred rewards obtained by these methods represent the global assessment of a whole or partial path but do not accumulate the rewards of local changes accurately, so the sparse global rewards may lead to non-convergence and unstable training performances. In this paper, we propose an online reward generation method guided by the online causation monitoring of STL. Our approach continuously monitors system behavior against an STL specification at each control step, computing the quantitative distance toward satisfaction or violation and thereby producing rewards that reflect instantaneous state dynamics. Additionally, we provide a smooth approximation of the causation semantics to overcome the discontinuity of the causation semantics and make it differentiable for using deep-RL methods. We have implemented a prototype tool and evaluated it in the Gym environment on a variety of continuously controlled benchmarks. Experimental results show that our proposed STL-guided RL method with online causation semantics outperforms existing relevant STL-guided RL methods, providing a more robust and efficient reward generation framework for deep-RL.
IrrMap: A Large-Scale Comprehensive Dataset for Irrigation Method Mapping
May 13, 2025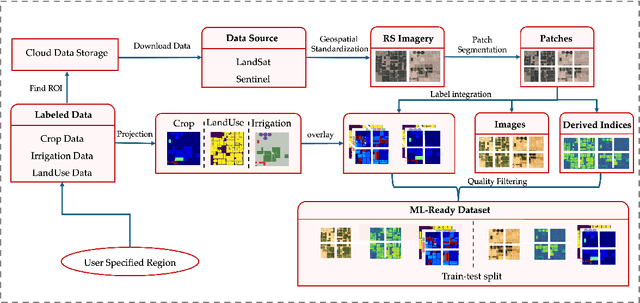
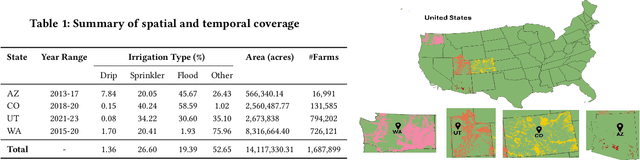

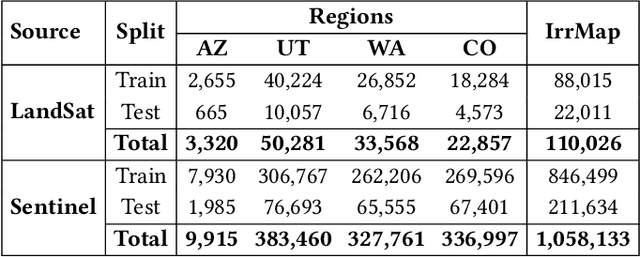
Abstract:We introduce IrrMap, the first large-scale dataset (1.1 million patches) for irrigation method mapping across regions. IrrMap consists of multi-resolution satellite imagery from LandSat and Sentinel, along with key auxiliary data such as crop type, land use, and vegetation indices. The dataset spans 1,687,899 farms and 14,117,330 acres across multiple western U.S. states from 2013 to 2023, providing a rich and diverse foundation for irrigation analysis and ensuring geospatial alignment and quality control. The dataset is ML-ready, with standardized 224x224 GeoTIFF patches, the multiple input modalities, carefully chosen train-test-split data, and accompanying dataloaders for seamless deep learning model training andbenchmarking in irrigation mapping. The dataset is also accompanied by a complete pipeline for dataset generation, enabling researchers to extend IrrMap to new regions for irrigation data collection or adapt it with minimal effort for other similar applications in agricultural and geospatial analysis. We also analyze the irrigation method distribution across crop groups, spatial irrigation patterns (using Shannon diversity indices), and irrigated area variations for both LandSat and Sentinel, providing insights into regional and resolution-based differences. To promote further exploration, we openly release IrrMap, along with the derived datasets, benchmark models, and pipeline code, through a GitHub repository: https://github.com/Nibir088/IrrMap and Data repository: https://huggingface.co/Nibir/IrrMap, providing comprehensive documentation and implementation details.
TPIE: Topology-Preserved Image Editing With Text Instructions
Nov 22, 2024



Abstract:Preserving topological structures is important in real-world applications, particularly in sensitive domains such as healthcare and medicine, where the correctness of human anatomy is critical. However, most existing image editing models focus on manipulating intensity and texture features, often overlooking object geometry within images. To address this issue, this paper introduces a novel method, Topology-Preserved Image Editing with text instructions (TPIE), that for the first time ensures the topology and geometry remaining intact in edited images through text-guided generative diffusion models. More specifically, our method treats newly generated samples as deformable variations of a given input template, allowing for controllable and structure-preserving edits. Our proposed TPIE framework consists of two key modules: (i) an autoencoder-based registration network that learns latent representations of object transformations, parameterized by velocity fields, from pairwise training images; and (ii) a novel latent conditional geometric diffusion (LCDG) model efficiently capturing the data distribution of learned transformation features conditioned on custom-defined text instructions. We validate TPIE on a diverse set of 2D and 3D images and compare them with state-of-the-art image editing approaches. Experimental results show that our method outperforms other baselines in generating more realistic images with well-preserved topology. Our code will be made publicly available on Github.
Invariant Shape Representation Learning For Image Classification
Nov 19, 2024



Abstract:Geometric shape features have been widely used as strong predictors for image classification. Nevertheless, most existing classifiers such as deep neural networks (DNNs) directly leverage the statistical correlations between these shape features and target variables. However, these correlations can often be spurious and unstable across different environments (e.g., in different age groups, certain types of brain changes have unstable relations with neurodegenerative disease); hence leading to biased or inaccurate predictions. In this paper, we introduce a novel framework that for the first time develops invariant shape representation learning (ISRL) to further strengthen the robustness of image classifiers. In contrast to existing approaches that mainly derive features in the image space, our model ISRL is designed to jointly capture invariant features in latent shape spaces parameterized by deformable transformations. To achieve this goal, we develop a new learning paradigm based on invariant risk minimization (IRM) to learn invariant representations of image and shape features across multiple training distributions/environments. By embedding the features that are invariant with regard to target variables in different environments, our model consistently offers more accurate predictions. We validate our method by performing classification tasks on both simulated 2D images, real 3D brain and cine cardiovascular magnetic resonance images (MRIs). Our code is publicly available at https://github.com/tonmoy-hossain/ISRL.
Learning Geodesics of Geometric Shape Deformations From Images
Oct 24, 2024Abstract:This paper presents a novel method, named geodesic deformable networks (GDN), that for the first time enables the learning of geodesic flows of deformation fields derived from images. In particular, the capability of our proposed GDN being able to predict geodesics is important for quantifying and comparing deformable shape presented in images. The geodesic deformations, also known as optimal transformations that align pairwise images, are often parameterized by a time sequence of smooth vector fields governed by nonlinear differential equations. A bountiful literature has been focusing on learning the initial conditions (e.g., initial velocity fields) based on registration networks. However, the definition of geodesics central to deformation-based shape analysis is blind to the networks. To address this problem, we carefully develop an efficient neural operator to treat the geodesics as unknown mapping functions learned from the latent deformation spaces. A composition of integral operators and smooth activation functions is then formulated to effectively approximate such mappings. In contrast to previous works, our GDN jointly optimizes a newly defined geodesic loss, which adds additional benefits to promote the network regularizability and generalizability. We demonstrate the effectiveness of GDN on both 2D synthetic data and 3D real brain magnetic resonance imaging (MRI).
Interior Object Geometry via Fitted Frames
Jul 19, 2024
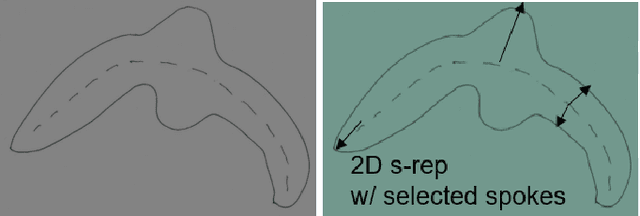

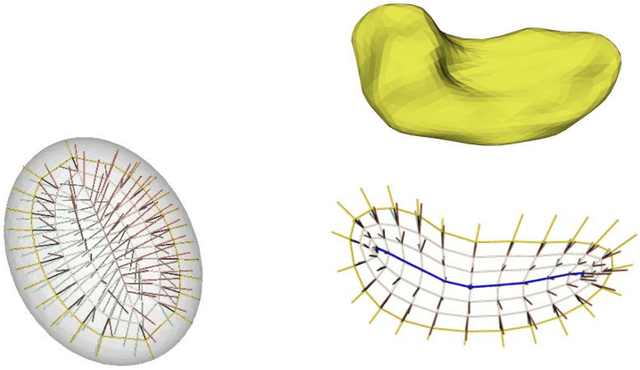
Abstract:We describe a representation targeted for anatomic objects which is designed to enable strong locational correspondence within object populations and thus to provide powerful object statistics. The method generates fitted frames on the boundary and in the interior of objects and produces alignment-free geometric features from them. It accomplishes this by understanding an object as the diffeomorphic deformation of an ellipsoid and using a skeletal representation fitted throughout the deformation to produce a model of the target object, where the object is provided initially in the form of a boundary mesh. Via classification performance on hippocampi shape between individuals with a disorder vs. others, we compare our method to two state-of-the-art methods for producing object representations that are intended to capture geometric correspondence across a population of objects and to yield geometric features useful for statistics, and we show improved classification performance by this new representation, which we call the evolutionary s-rep. The geometric features that are derived from each of the representations, especially via fitted frames, is discussed.
TLRN: Temporal Latent Residual Networks For Large Deformation Image Registration
Jul 15, 2024



Abstract:This paper presents a novel approach, termed {\em Temporal Latent Residual Network (TLRN)}, to predict a sequence of deformation fields in time-series image registration. The challenge of registering time-series images often lies in the occurrence of large motions, especially when images differ significantly from a reference (e.g., the start of a cardiac cycle compared to the peak stretching phase). To achieve accurate and robust registration results, we leverage the nature of motion continuity and exploit the temporal smoothness in consecutive image frames. Our proposed TLRN highlights a temporal residual network with residual blocks carefully designed in latent deformation spaces, which are parameterized by time-sequential initial velocity fields. We treat a sequence of residual blocks over time as a dynamic training system, where each block is designed to learn the residual function between desired deformation features and current input accumulated from previous time frames. We validate the effectivenss of TLRN on both synthetic data and real-world cine cardiac magnetic resonance (CMR) image videos. Our experimental results shows that TLRN is able to achieve substantially improved registration accuracy compared to the state-of-the-art. Our code is publicly available at https://github.com/nellie689/TLRN.
LaMoD: Latent Motion Diffusion Model For Myocardial Strain Generation
Jul 02, 2024



Abstract:Motion and deformation analysis of cardiac magnetic resonance (CMR) imaging videos is crucial for assessing myocardial strain of patients with abnormal heart functions. Recent advances in deep learning-based image registration algorithms have shown promising results in predicting motion fields from routinely acquired CMR sequences. However, their accuracy often diminishes in regions with subtle appearance change, with errors propagating over time. Advanced imaging techniques, such as displacement encoding with stimulated echoes (DENSE) CMR, offer highly accurate and reproducible motion data but require additional image acquisition, which poses challenges in busy clinical flows. In this paper, we introduce a novel Latent Motion Diffusion model (LaMoD) to predict highly accurate DENSE motions from standard CMR videos. More specifically, our method first employs an encoder from a pre-trained registration network that learns latent motion features (also considered as deformation-based shape features) from image sequences. Supervised by the ground-truth motion provided by DENSE, LaMoD then leverages a probabilistic latent diffusion model to reconstruct accurate motion from these extracted features. Experimental results demonstrate that our proposed method, LaMoD, significantly improves the accuracy of motion analysis in standard CMR images; hence improving myocardial strain analysis in clinical settings for cardiac patients. Our code will be publicly available on upon acceptance.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge