Jiarui Xing
PathoSyn: Imaging-Pathology MRI Synthesis via Disentangled Deviation Diffusion
Dec 29, 2025Abstract:We present PathoSyn, a unified generative framework for Magnetic Resonance Imaging (MRI) image synthesis that reformulates imaging-pathology as a disentangled additive deviation on a stable anatomical manifold. Current generative models typically operate in the global pixel domain or rely on binary masks, these paradigms often suffer from feature entanglement, leading to corrupted anatomical substrates or structural discontinuities. PathoSyn addresses these limitations by decomposing the synthesis task into deterministic anatomical reconstruction and stochastic deviation modeling. Central to our framework is a Deviation-Space Diffusion Model designed to learn the conditional distribution of pathological residuals, thereby capturing localized intensity variations while preserving global structural integrity by construction. To ensure spatial coherence, the diffusion process is coupled with a seam-aware fusion strategy and an inference-time stabilization module, which collectively suppress boundary artifacts and produce high-fidelity internal lesion heterogeneity. PathoSyn provides a mathematically principled pipeline for generating high-fidelity patient-specific synthetic datasets, facilitating the development of robust diagnostic algorithms in low-data regimes. By allowing interpretable counterfactual disease progression modeling, the framework supports precision intervention planning and provides a controlled environment for benchmarking clinical decision-support systems. Quantitative and qualitative evaluations on tumor imaging benchmarks demonstrate that PathoSyn significantly outperforms holistic diffusion and mask-conditioned baselines in both perceptual realism and anatomical fidelity. The source code of this work will be made publicly available.
TLRN: Temporal Latent Residual Networks For Large Deformation Image Registration
Jul 15, 2024



Abstract:This paper presents a novel approach, termed {\em Temporal Latent Residual Network (TLRN)}, to predict a sequence of deformation fields in time-series image registration. The challenge of registering time-series images often lies in the occurrence of large motions, especially when images differ significantly from a reference (e.g., the start of a cardiac cycle compared to the peak stretching phase). To achieve accurate and robust registration results, we leverage the nature of motion continuity and exploit the temporal smoothness in consecutive image frames. Our proposed TLRN highlights a temporal residual network with residual blocks carefully designed in latent deformation spaces, which are parameterized by time-sequential initial velocity fields. We treat a sequence of residual blocks over time as a dynamic training system, where each block is designed to learn the residual function between desired deformation features and current input accumulated from previous time frames. We validate the effectivenss of TLRN on both synthetic data and real-world cine cardiac magnetic resonance (CMR) image videos. Our experimental results shows that TLRN is able to achieve substantially improved registration accuracy compared to the state-of-the-art. Our code is publicly available at https://github.com/nellie689/TLRN.
LaMoD: Latent Motion Diffusion Model For Myocardial Strain Generation
Jul 02, 2024



Abstract:Motion and deformation analysis of cardiac magnetic resonance (CMR) imaging videos is crucial for assessing myocardial strain of patients with abnormal heart functions. Recent advances in deep learning-based image registration algorithms have shown promising results in predicting motion fields from routinely acquired CMR sequences. However, their accuracy often diminishes in regions with subtle appearance change, with errors propagating over time. Advanced imaging techniques, such as displacement encoding with stimulated echoes (DENSE) CMR, offer highly accurate and reproducible motion data but require additional image acquisition, which poses challenges in busy clinical flows. In this paper, we introduce a novel Latent Motion Diffusion model (LaMoD) to predict highly accurate DENSE motions from standard CMR videos. More specifically, our method first employs an encoder from a pre-trained registration network that learns latent motion features (also considered as deformation-based shape features) from image sequences. Supervised by the ground-truth motion provided by DENSE, LaMoD then leverages a probabilistic latent diffusion model to reconstruct accurate motion from these extracted features. Experimental results demonstrate that our proposed method, LaMoD, significantly improves the accuracy of motion analysis in standard CMR images; hence improving myocardial strain analysis in clinical settings for cardiac patients. Our code will be publicly available on upon acceptance.
Multimodal Learning To Improve Cardiac Late Mechanical Activation Detection From Cine MR Images
Feb 28, 2024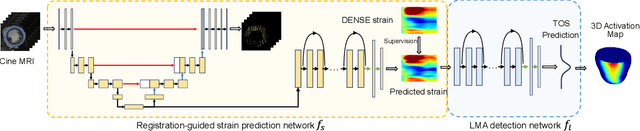
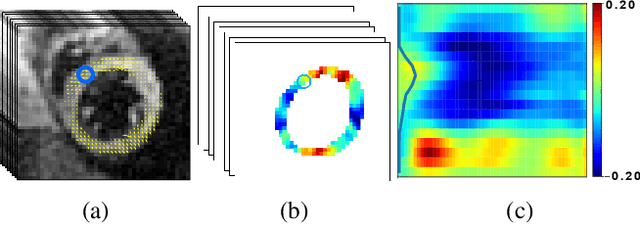
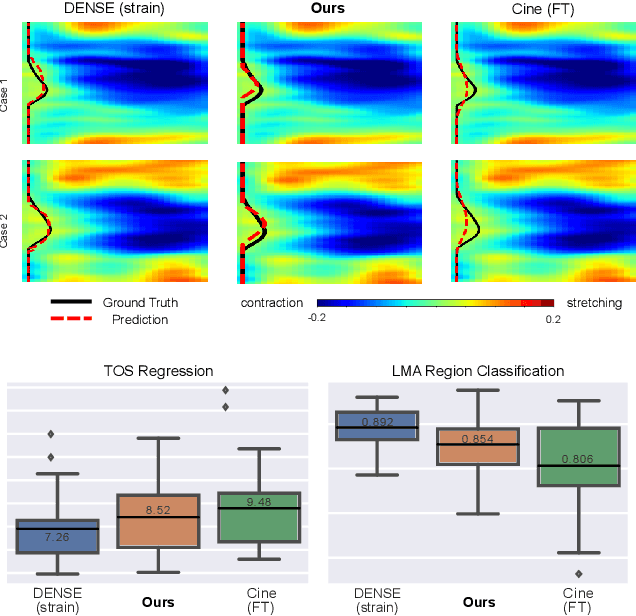
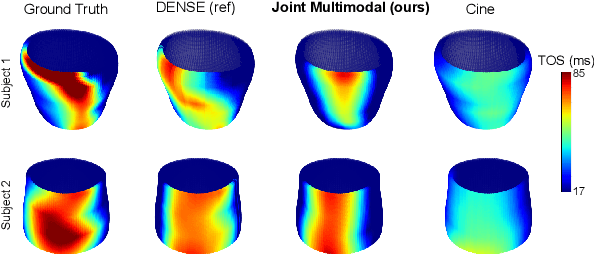
Abstract:This paper presents a multimodal deep learning framework that utilizes advanced image techniques to improve the performance of clinical analysis heavily dependent on routinely acquired standard images. More specifically, we develop a joint learning network that for the first time leverages the accuracy and reproducibility of myocardial strains obtained from Displacement Encoding with Stimulated Echo (DENSE) to guide the analysis of cine cardiac magnetic resonance (CMR) imaging in late mechanical activation (LMA) detection. An image registration network is utilized to acquire the knowledge of cardiac motions, an important feature estimator of strain values, from standard cine CMRs. Our framework consists of two major components: (i) a DENSE-supervised strain network leveraging latent motion features learned from a registration network to predict myocardial strains; and (ii) a LMA network taking advantage of the predicted strain for effective LMA detection. Experimental results show that our proposed work substantially improves the performance of strain analysis and LMA detection from cine CMR images, aligning more closely with the achievements of DENSE.
MetaMorph: Learning Metamorphic Image Transformation With Appearance Changes
Mar 08, 2023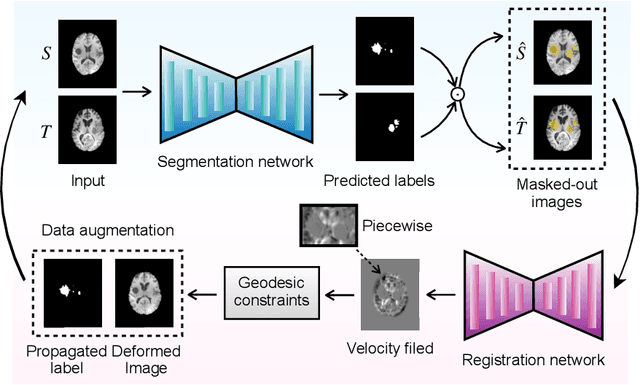
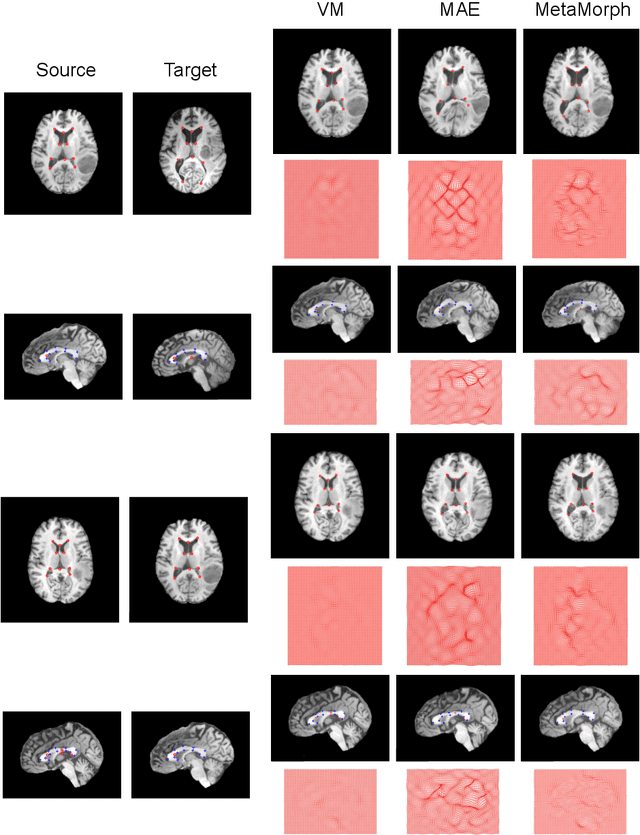
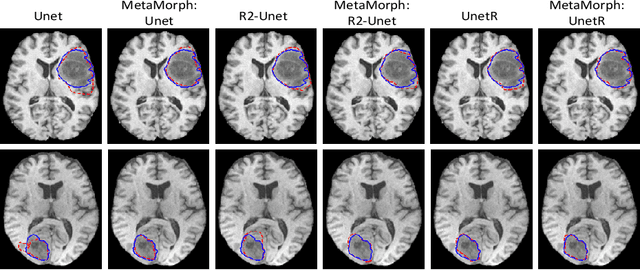
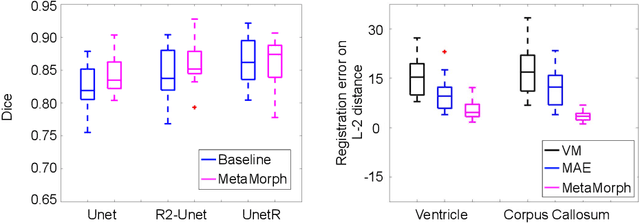
Abstract:This paper presents a novel predictive model, MetaMorph, for metamorphic registration of images with appearance changes (i.e., caused by brain tumors). In contrast to previous learning-based registration methods that have little or no control over appearance-changes, our model introduces a new regularization that can effectively suppress the negative effects of appearance changing areas. In particular, we develop a piecewise regularization on the tangent space of diffeomorphic transformations (also known as initial velocity fields) via learned segmentation maps of abnormal regions. The geometric transformation and appearance changes are treated as joint tasks that are mutually beneficial. Our model MetaMorph is more robust and accurate when searching for an optimal registration solution under the guidance of segmentation, which in turn improves the segmentation performance by providing appropriately augmented training labels. We validate MetaMorph on real 3D human brain tumor magnetic resonance imaging (MRI) scans. Experimental results show that our model outperforms the state-of-the-art learning-based registration models. The proposed MetaMorph has great potential in various image-guided clinical interventions, e.g., real-time image-guided navigation systems for tumor removal surgery.
Multitask Learning for Improved Late Mechanical Activation Detection of Heart from Cine DENSE MRI
Nov 11, 2022



Abstract:The selection of an optimal pacing site, which is ideally scar-free and late activated, is critical to the response of cardiac resynchronization therapy (CRT). Despite the success of current approaches formulating the detection of such late mechanical activation (LMA) regions as a problem of activation time regression, their accuracy remains unsatisfactory, particularly in cases where myocardial scar exists. To address this issue, this paper introduces a multi-task deep learning framework that simultaneously estimates LMA amount and classify the scar-free LMA regions based on cine displacement encoding with stimulated echoes (DENSE) magnetic resonance imaging (MRI). With a newly introduced auxiliary LMA region classification sub-network, our proposed model shows more robustness to the complex pattern cause by myocardial scar, significantly eliminates their negative effects in LMA detection, and in turn improves the performance of scar classification. To evaluate the effectiveness of our method, we tests our model on real cardiac MR images and compare the predicted LMA with the state-of-the-art approaches. It shows that our approach achieves substantially increased accuracy. In addition, we employ the gradient-weighted class activation mapping (Grad-CAM) to visualize the feature maps learned by all methods. Experimental results suggest that our proposed model better recognizes the LMA region pattern.
Joint Deep Learning for Improved Myocardial Scar Detection from Cardiac MRI
Nov 11, 2022



Abstract:Automated identification of myocardial scar from late gadolinium enhancement cardiac magnetic resonance images (LGE-CMR) is limited by image noise and artifacts such as those related to motion and partial volume effect. This paper presents a novel joint deep learning (JDL) framework that improves such tasks by utilizing simultaneously learned myocardium segmentations to eliminate negative effects from non-region-of-interest areas. In contrast to previous approaches treating scar detection and myocardium segmentation as separate or parallel tasks, our proposed method introduces a message passing module where the information of myocardium segmentation is directly passed to guide scar detectors. This newly designed network will efficiently exploit joint information from the two related tasks and use all available sources of myocardium segmentation to benefit scar identification. We demonstrate the effectiveness of JDL on LGE-CMR images for automated left ventricular (LV) scar detection, with great potential to improve risk prediction in patients with both ischemic and non-ischemic heart disease and to improve response rates to cardiac resynchronization therapy (CRT) for heart failure patients. Experimental results show that our proposed approach outperforms multiple state-of-the-art methods, including commonly used two-step segmentation-classification networks, and multitask learning schemes where subtasks are indirectly interacted.
Mixture Probabilistic Principal Geodesic Analysis
Sep 05, 2019



Abstract:Dimensionality reduction on Riemannian manifolds is challenging due to the complex nonlinear data structures. While probabilistic principal geodesic analysis~(PPGA) has been proposed to generalize conventional principal component analysis (PCA) onto manifolds, its effectiveness is limited to data with a single modality. In this paper, we present a novel Gaussian latent variable model that provides a unique way to integrate multiple PGA models into a maximum-likelihood framework. This leads to a well-defined mixture model of probabilistic principal geodesic analysis (MPPGA) on sub-populations, where parameters of the principal subspaces are automatically estimated by employing an Expectation Maximization algorithm. We further develop a mixture Bayesian PGA (MBPGA) model that automatically reduces data dimensionality by suppressing irrelevant principal geodesics. We demonstrate the advantages of our model in the contexts of clustering and statistical shape analysis, using synthetic sphere data, real corpus callosum, and mandible data from human brain magnetic resonance~(MR) and CT images.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge