Nian Wu
Robust Orthogonal NMF with Label Propagation for Image Clustering
Apr 30, 2025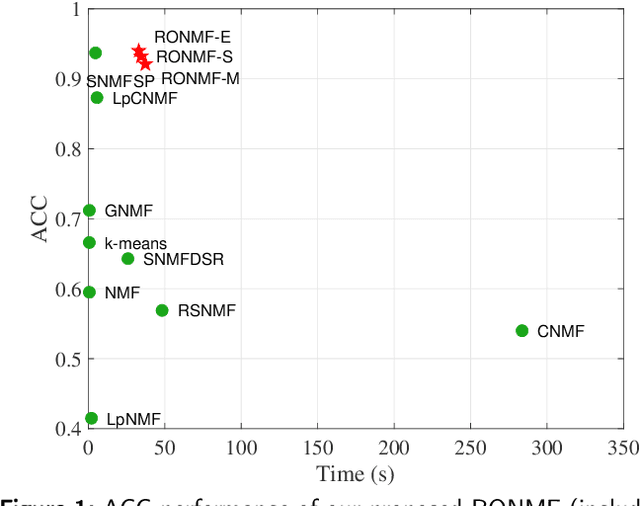
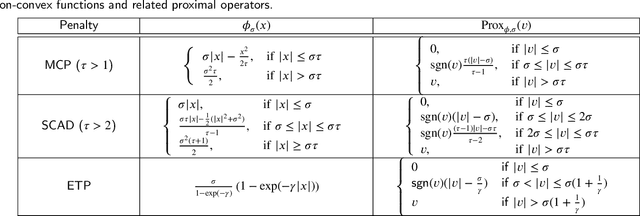
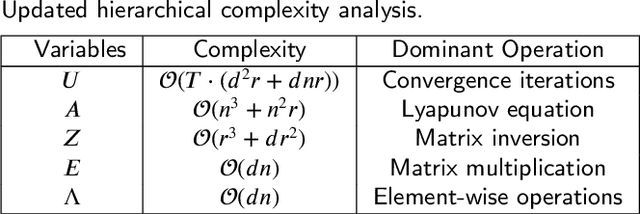
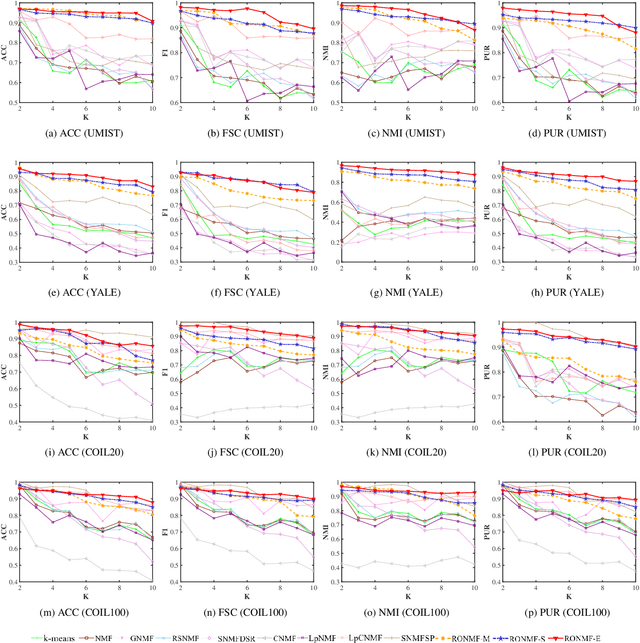
Abstract:Non-negative matrix factorization (NMF) is a popular unsupervised learning approach widely used in image clustering. However, in real-world clustering scenarios, most existing NMF methods are highly sensitive to noise corruption and are unable to effectively leverage limited supervised information. To overcome these drawbacks, we propose a unified non-convex framework with label propagation called robust orthogonal nonnegative matrix factorization (RONMF). This method not only considers the graph Laplacian and label propagation as regularization terms but also introduces a more effective non-convex structure to measure the reconstruction error and imposes orthogonal constraints on the basis matrix to reduce the noise corruption, thereby achieving higher robustness. To solve RONMF, we develop an alternating direction method of multipliers (ADMM)-based optimization algorithm. In particular, all subproblems have closed-form solutions, which ensures its efficiency. Experimental evaluations on eight public image datasets demonstrate that the proposed RONMF outperforms state-of-the-art NMF methods across various standard metrics and shows excellent robustness. The code will be available at https://github.com/slinda-liu.
Learning Geodesics of Geometric Shape Deformations From Images
Oct 24, 2024Abstract:This paper presents a novel method, named geodesic deformable networks (GDN), that for the first time enables the learning of geodesic flows of deformation fields derived from images. In particular, the capability of our proposed GDN being able to predict geodesics is important for quantifying and comparing deformable shape presented in images. The geodesic deformations, also known as optimal transformations that align pairwise images, are often parameterized by a time sequence of smooth vector fields governed by nonlinear differential equations. A bountiful literature has been focusing on learning the initial conditions (e.g., initial velocity fields) based on registration networks. However, the definition of geodesics central to deformation-based shape analysis is blind to the networks. To address this problem, we carefully develop an efficient neural operator to treat the geodesics as unknown mapping functions learned from the latent deformation spaces. A composition of integral operators and smooth activation functions is then formulated to effectively approximate such mappings. In contrast to previous works, our GDN jointly optimizes a newly defined geodesic loss, which adds additional benefits to promote the network regularizability and generalizability. We demonstrate the effectiveness of GDN on both 2D synthetic data and 3D real brain magnetic resonance imaging (MRI).
TLRN: Temporal Latent Residual Networks For Large Deformation Image Registration
Jul 15, 2024



Abstract:This paper presents a novel approach, termed {\em Temporal Latent Residual Network (TLRN)}, to predict a sequence of deformation fields in time-series image registration. The challenge of registering time-series images often lies in the occurrence of large motions, especially when images differ significantly from a reference (e.g., the start of a cardiac cycle compared to the peak stretching phase). To achieve accurate and robust registration results, we leverage the nature of motion continuity and exploit the temporal smoothness in consecutive image frames. Our proposed TLRN highlights a temporal residual network with residual blocks carefully designed in latent deformation spaces, which are parameterized by time-sequential initial velocity fields. We treat a sequence of residual blocks over time as a dynamic training system, where each block is designed to learn the residual function between desired deformation features and current input accumulated from previous time frames. We validate the effectivenss of TLRN on both synthetic data and real-world cine cardiac magnetic resonance (CMR) image videos. Our experimental results shows that TLRN is able to achieve substantially improved registration accuracy compared to the state-of-the-art. Our code is publicly available at https://github.com/nellie689/TLRN.
LaMoD: Latent Motion Diffusion Model For Myocardial Strain Generation
Jul 02, 2024



Abstract:Motion and deformation analysis of cardiac magnetic resonance (CMR) imaging videos is crucial for assessing myocardial strain of patients with abnormal heart functions. Recent advances in deep learning-based image registration algorithms have shown promising results in predicting motion fields from routinely acquired CMR sequences. However, their accuracy often diminishes in regions with subtle appearance change, with errors propagating over time. Advanced imaging techniques, such as displacement encoding with stimulated echoes (DENSE) CMR, offer highly accurate and reproducible motion data but require additional image acquisition, which poses challenges in busy clinical flows. In this paper, we introduce a novel Latent Motion Diffusion model (LaMoD) to predict highly accurate DENSE motions from standard CMR videos. More specifically, our method first employs an encoder from a pre-trained registration network that learns latent motion features (also considered as deformation-based shape features) from image sequences. Supervised by the ground-truth motion provided by DENSE, LaMoD then leverages a probabilistic latent diffusion model to reconstruct accurate motion from these extracted features. Experimental results demonstrate that our proposed method, LaMoD, significantly improves the accuracy of motion analysis in standard CMR images; hence improving myocardial strain analysis in clinical settings for cardiac patients. Our code will be publicly available on upon acceptance.
Multimodal Learning To Improve Cardiac Late Mechanical Activation Detection From Cine MR Images
Feb 28, 2024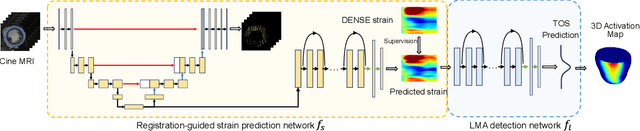
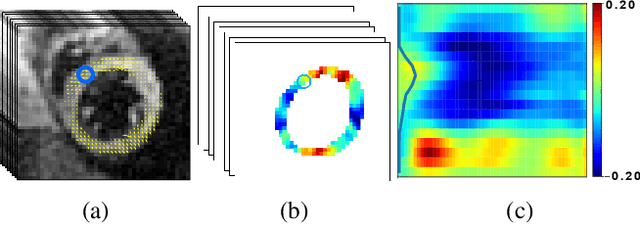
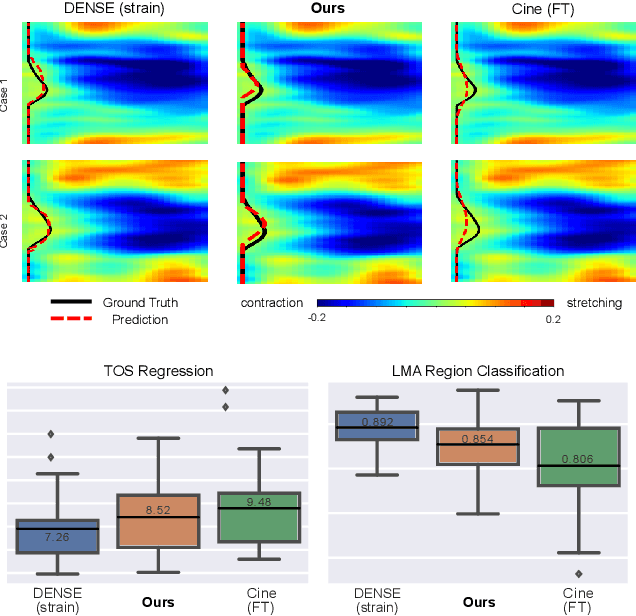
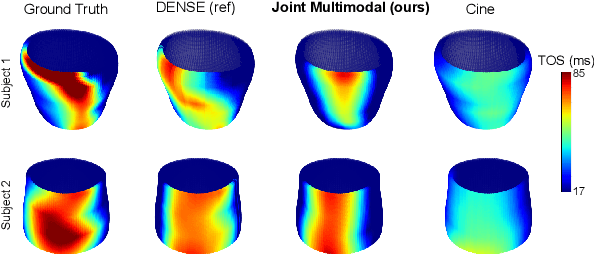
Abstract:This paper presents a multimodal deep learning framework that utilizes advanced image techniques to improve the performance of clinical analysis heavily dependent on routinely acquired standard images. More specifically, we develop a joint learning network that for the first time leverages the accuracy and reproducibility of myocardial strains obtained from Displacement Encoding with Stimulated Echo (DENSE) to guide the analysis of cine cardiac magnetic resonance (CMR) imaging in late mechanical activation (LMA) detection. An image registration network is utilized to acquire the knowledge of cardiac motions, an important feature estimator of strain values, from standard cine CMRs. Our framework consists of two major components: (i) a DENSE-supervised strain network leveraging latent motion features learned from a registration network to predict myocardial strains; and (ii) a LMA network taking advantage of the predicted strain for effective LMA detection. Experimental results show that our proposed work substantially improves the performance of strain analysis and LMA detection from cine CMR images, aligning more closely with the achievements of DENSE.
NeurEPDiff: Neural Operators to Predict Geodesics in Deformation Spaces
Mar 13, 2023



Abstract:This paper presents NeurEPDiff, a novel network to fast predict the geodesics in deformation spaces generated by a well known Euler-Poincar\'e differential equation (EPDiff). To achieve this, we develop a neural operator that for the first time learns the evolving trajectory of geodesic deformations parameterized in the tangent space of diffeomorphisms(a.k.a velocity fields). In contrast to previous methods that purely fit the training images, our proposed NeurEPDiff learns a nonlinear mapping function between the time-dependent velocity fields. A composition of integral operators and smooth activation functions is formulated in each layer of NeurEPDiff to effectively approximate such mappings. The fact that NeurEPDiff is able to rapidly provide the numerical solution of EPDiff (given any initial condition) results in a significantly reduced computational cost of geodesic shooting of diffeomorphisms in a high-dimensional image space. Additionally, the properties of discretiztion/resolution-invariant of NeurEPDiff make its performance generalizable to multiple image resolutions after being trained offline. We demonstrate the effectiveness of NeurEPDiff in registering two image datasets: 2D synthetic data and 3D brain resonance imaging (MRI). The registration accuracy and computational efficiency are compared with the state-of-the-art diffeomophic registration algorithms with geodesic shooting.
Predicting the protein-ligand affinity from molecular dynamics trajectories
Aug 19, 2022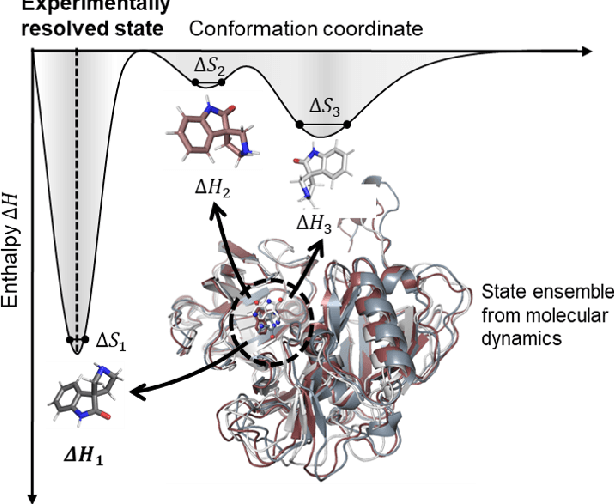
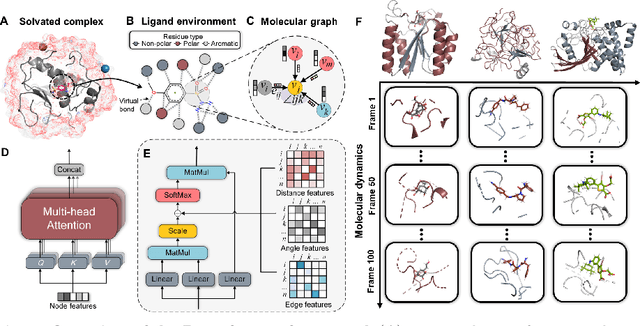
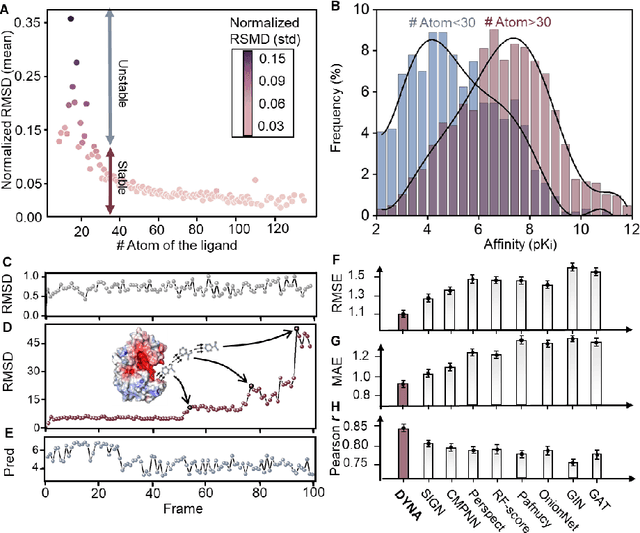
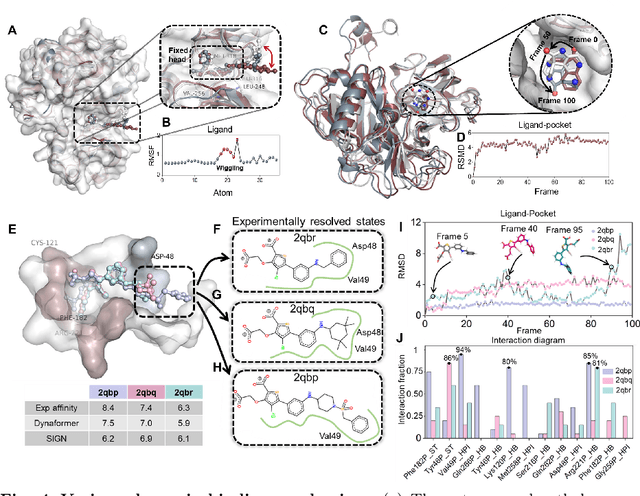
Abstract:The accurate protein-ligand binding affinity prediction is essential in drug design and many other molecular recognition problems. Despite many advances in affinity prediction based on machine learning techniques, they are still limited since the protein-ligand binding is determined by the dynamics of atoms and molecules. To this end, we curated an MD dataset containing 3,218 dynamic protein-ligand complexes and further developed Dynaformer, a graph-based deep learning framework. Dynaformer can fully capture the dynamic binding rules by considering various geometric characteristics of the interaction. Our method shows superior performance over the methods hitherto reported. Moreover, we performed virtual screening on heat shock protein 90 (HSP90) by integrating our model with structure-based docking. We benchmarked our performance against other baselines, demonstrating that our method can identify the molecule with the highest experimental potency. We anticipate that large-scale MD dataset and machine learning models will form a new synergy, providing a new route towards accelerated drug discovery and optimization.
Hybrid Atlas Building with Deep Registration Priors
Dec 13, 2021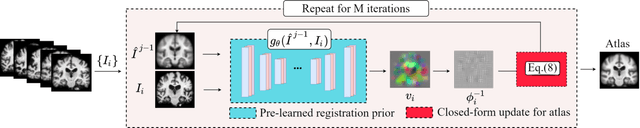
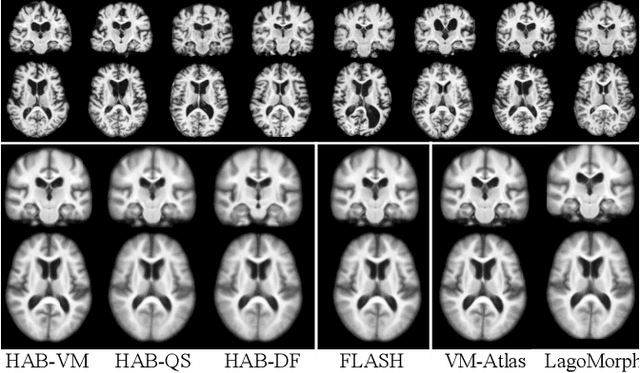
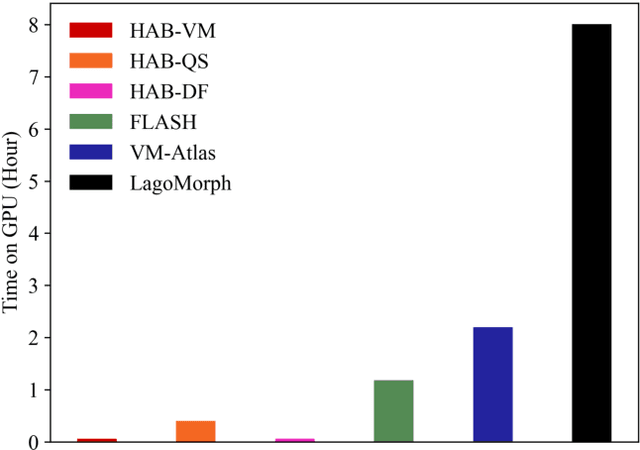
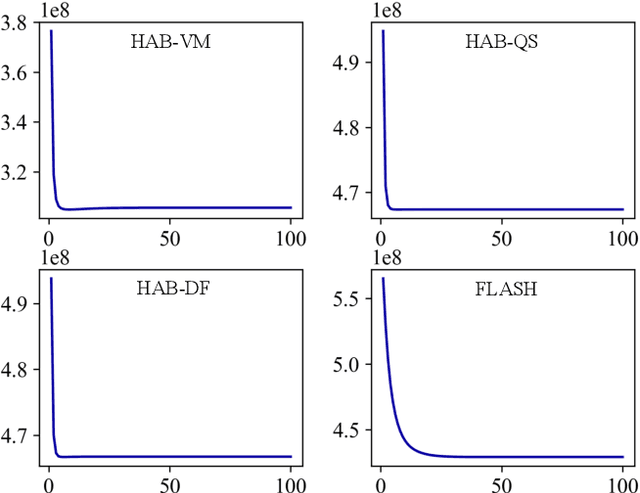
Abstract:Registration-based atlas building often poses computational challenges in high-dimensional image spaces. In this paper, we introduce a novel hybrid atlas building algorithm that fast estimates atlas from large-scale image datasets with much reduced computational cost. In contrast to previous approaches that iteratively perform registration tasks between an estimated atlas and individual images, we propose to use learned priors of registration from pre-trained neural networks. This newly developed hybrid framework features several advantages of (i) providing an efficient way of atlas building without losing the quality of results, and (ii) offering flexibility in utilizing a wide variety of deep learning based registration methods. We demonstrate the effectiveness of this proposed model on 3D brain magnetic resonance imaging (MRI) scans.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge