Matthew Toews
The Mechanics of CNN Filtering with Rectification
Dec 30, 2025Abstract:This paper proposes elementary information mechanics as a new model for understanding the mechanical properties of convolutional filtering with rectification, inspired by physical theories of special relativity and quantum mechanics. We consider kernels decomposed into orthogonal even and odd components. Even components cause image content to diffuse isotropically while preserving the center of mass, analogously to rest or potential energy with zero net momentum. Odd kernels cause directional displacement of the center of mass, analogously to kinetic energy with non-zero momentum. The speed of information displacement is linearly related to the ratio of odd vs total kernel energy. Even-Odd properties are analyzed in the spectral domain via the discrete cosine transform (DCT), where the structure of small convolutional filters (e.g. $3 \times 3$ pixels) is dominated by low-frequency bases, specifically the DC $Σ$ and gradient components $\nabla$, which define the fundamental modes of information propagation. To our knowledge, this is the first work demonstrating the link between information processing in generic CNNs and the energy-momentum relation, a cornerstone of modern relativistic physics.
NTIRE 2025 Challenge on HR Depth from Images of Specular and Transparent Surfaces
Jun 06, 2025Abstract:This paper reports on the NTIRE 2025 challenge on HR Depth From images of Specular and Transparent surfaces, held in conjunction with the New Trends in Image Restoration and Enhancement (NTIRE) workshop at CVPR 2025. This challenge aims to advance the research on depth estimation, specifically to address two of the main open issues in the field: high-resolution and non-Lambertian surfaces. The challenge proposes two tracks on stereo and single-image depth estimation, attracting about 177 registered participants. In the final testing stage, 4 and 4 participating teams submitted their models and fact sheets for the two tracks.
Neural Architecture Search by Learning a Hierarchical Search Space
Mar 27, 2025Abstract:Monte-Carlo Tree Search (MCTS) is a powerful tool for many non-differentiable search related problems such as adversarial games. However, the performance of such approach highly depends on the order of the nodes that are considered at each branching of the tree. If the first branches cannot distinguish between promising and deceiving configurations for the final task, the efficiency of the search is exponentially reduced. In Neural Architecture Search (NAS), as only the final architecture matters, the visiting order of the branching can be optimized to improve learning. In this paper, we study the application of MCTS to NAS for image classification. We analyze several sampling methods and branching alternatives for MCTS and propose to learn the branching by hierarchical clustering of architectures based on their similarity. The similarity is measured by the pairwise distance of output vectors of architectures. Extensive experiments on two challenging benchmarks on CIFAR10 and ImageNet show that MCTS, if provided with a good branching hierarchy, can yield promising solutions more efficiently than other approaches for NAS problems.
Coloring Deep CNN Layers with Activation Hue Loss
Oct 05, 2023Abstract:This paper proposes a novel hue-like angular parameter to model the structure of deep convolutional neural network (CNN) activation space, referred to as the {\em activation hue}, for the purpose of regularizing models for more effective learning. The activation hue generalizes the notion of color hue angle in standard 3-channel RGB intensity space to $N$-channel activation space. A series of observations based on nearest neighbor indexing of activation vectors with pre-trained networks indicate that class-informative activations are concentrated about an angle $\theta$ in both the $(x,y)$ image plane and in multi-channel activation space. A regularization term in the form of hue-like angular $\theta$ labels is proposed to complement standard one-hot loss. Training from scratch using combined one-hot + activation hue loss improves classification performance modestly for a wide variety of classification tasks, including ImageNet.
Balanced Mixture of SuperNets for Learning the CNN Pooling Architecture
Jun 21, 2023



Abstract:Downsampling layers, including pooling and strided convolutions, are crucial components of the convolutional neural network architecture that determine both the granularity/scale of image feature analysis as well as the receptive field size of a given layer. To fully understand this problem, we analyse the performance of models independently trained with each pooling configurations on CIFAR10, using a ResNet20 network, and show that the position of the downsampling layers can highly influence the performance of a network and predefined downsampling configurations are not optimal. Network Architecture Search (NAS) might be used to optimize downsampling configurations as an hyperparameter. However, we find that common one-shot NAS based on a single SuperNet does not work for this problem. We argue that this is because a SuperNet trained for finding the optimal pooling configuration fully shares its parameters among all pooling configurations. This makes its training hard, because learning some configurations can harm the performance of others. Therefore, we propose a balanced mixture of SuperNets that automatically associates pooling configurations to different weight models and helps to reduce the weight-sharing and inter-influence of pooling configurations on the SuperNet parameters. We evaluate our proposed approach on CIFAR10, CIFAR100, as well as Food101 and show that in all cases, our model outperforms other approaches and improves over the default pooling configurations.
PSDNet: Determination of Particle Size Distributions Using Synthetic Soil Images and Convolutional Neural Networks
Mar 07, 2023



Abstract:This project aimed to determine the grain size distribution of granular materials from images using convolutional neural networks. The application of ConvNet and pretrained ConvNet models, including AlexNet, SqueezeNet, GoogLeNet, InceptionV3, DenseNet201, MobileNetV2, ResNet18, ResNet50, ResNet101, Xception, InceptionResNetV2, ShuffleNet, and NASNetMobile was studied. Synthetic images of granular materials created with the discrete element code YADE were used. All the models were trained and verified with grayscale and color band datasets with image sizes ranging from 32 to 160 pixels. The proposed ConvNet model predicts the percentages of mass retained on the finest sieve, coarsest sieve, and all sieves with root-mean-square errors of 1.8 %, 3.3 %, and 2.8 %, respectively, and a coefficient of determination of 0.99. For pretrained networks, root-mean-square errors of 2.4 % and 2.8 % were obtained for the finest sieve with feature extraction and transfer learning models, respectively.
U Symmetry-breaking Observed in Generic CNN Bottleneck Layers
Jun 05, 2022



Abstract:We report on a significant discovery linking deep convolutional neural networks (CNN) to biological vision and fundamental particle physics. A model of information propagation in a CNN is proposed via an analogy to an optical system, where bosonic particles (i.e. photons) are concentrated as the 2D spatial resolution of the image collapses to a focal point $1\times 1=1$. A 3D space $(x,y,t)$ is defined by $(x,y)$ coordinates in the image plane and CNN layer $t$, where a principal ray $(0,0,t)$ runs in the direction of information propagation through both the optical axis and the image center pixel located at $(x,y)=(0,0)$, about which the sharpest possible spatial focus is limited to a circle of confusion in the image plane. Our novel insight is to model the principal optical ray $(0,0,t)$ as geometrically equivalent to the medial vector in the positive orthant $I(x,y) \in R^{N+}$ of a $N$-channel activation space, e.g. along the greyscale (or luminance) vector $(t,t,t)$ in $RGB$ colour space. Information is thus concentrated into an energy potential $E(x,y,t)=\|I(x,y,t)\|^2$, which, particularly for bottleneck layers $t$ of generic CNNs, is highly concentrated and symmetric about the spatial origin $(0,0,t)$ and exhibits the well-known "Sombrero" potential of the boson particle. This symmetry is broken in classification, where bottleneck layers of generic pre-trained CNN models exhibit a consistent class-specific bias towards an angle $\theta \in U(1)$ defined simultaneously in the image plane and in activation feature space. Initial observations validate our hypothesis from generic pre-trained CNN activation maps and a bare-bones memory-based classification scheme, with no training or tuning. Training from scratch using a random $U(1)$ class label the leads to improved classification in all cases.
Registering Image Volumes using 3D SIFT and Discrete SP-Symmetry
May 30, 2022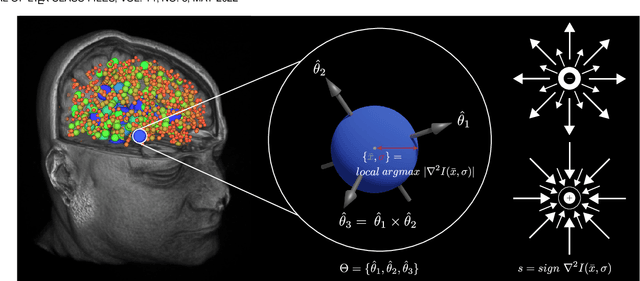
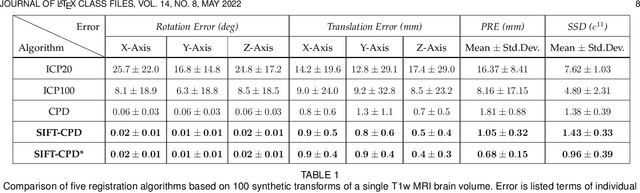
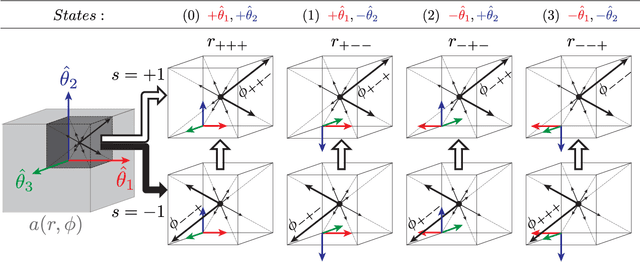
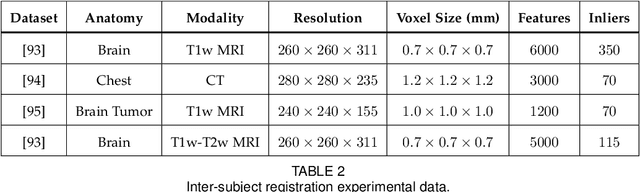
Abstract:This paper proposes to extend local image features in 3D to include invariance to discrete symmetry including inversion of spatial axes and image contrast. A binary feature sign $s \in \{-1,+1\}$ is defined as the sign of the Laplacian operator $\nabla^2$, and used to obtain a descriptor that is invariant to image sign inversion $s \rightarrow -s$ and 3D parity transforms $(x,y,z)\rightarrow(-x,-y,-z)$, i.e. SP-invariant or SP-symmetric. SP-symmetry applies to arbitrary scalar image fields $I: R^3 \rightarrow R^1$ mapping 3D coordinates $(x,y,z) \in R^3$ to scalar intensity $I(x,y,z) \in R^1$, generalizing the well-known charge conjugation and parity symmetry (CP-symmetry) applying to elementary charged particles. Feature orientation is modeled as a set of discrete states corresponding to potential axis reflections, independently of image contrast inversion. Two primary axis vectors are derived from image observations and potentially subject to reflection, and a third axis is an axial vector defined by the right-hand rule. Augmenting local feature properties with sign in addition to standard (location, scale, orientation) geometry leads to descriptors that are invariant to coordinate reflections and intensity contrast inversion. Feature properties are factored in to probabilistic point-based registration as symmetric kernels, based on a model of binary feature correspondence. Experiments using the well-known coherent point drift (CPD) algorithm demonstrate that SIFT-CPD kernels achieve the most accurate and rapid registration of the human brain and CT chest, including multiple MRI modalities of differing intensity contrast, and abnormal local variations such as tumors or occlusions. SIFT-CPD image registration is invariant to global scaling, rotation and translation and image intensity inversions of the input data.
GPU optimization of the 3D Scale-invariant Feature Transform Algorithm and a Novel BRIEF-inspired 3D Fast Descriptor
Dec 19, 2021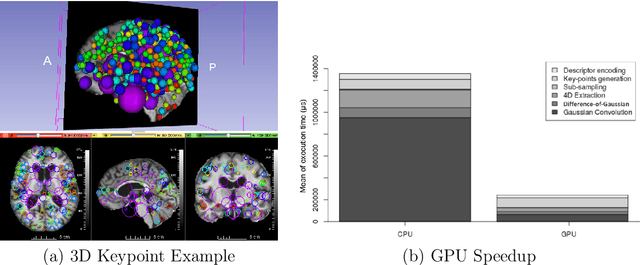
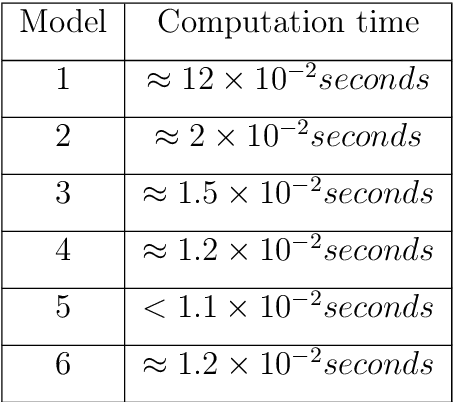
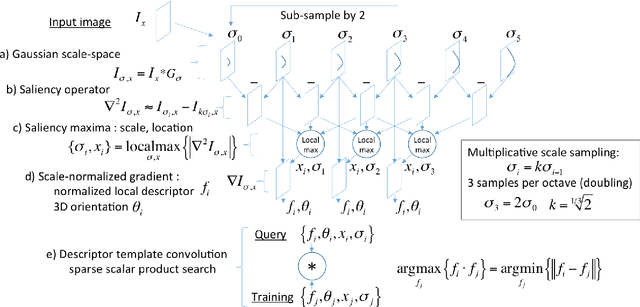
Abstract:This work details a highly efficient implementation of the 3D scale-invariant feature transform (SIFT) algorithm, for the purpose of machine learning from large sets of volumetric medical image data. The primary operations of the 3D SIFT code are implemented on a graphics processing unit (GPU), including convolution, sub-sampling, and 4D peak detection from scale-space pyramids. The performance improvements are quantified in keypoint detection and image-to-image matching experiments, using 3D MRI human brain volumes of different people. Computationally efficient 3D keypoint descriptors are proposed based on the Binary Robust Independent Elementary Feature (BRIEF) code, including a novel descriptor we call Ranked Robust Independent Elementary Features (RRIEF), and compared to the original 3D SIFT-Rank method\citep{toews2013efficient}. The GPU implementation affords a speedup of approximately 7X beyond an optimised CPU implementation, where computation time is reduced from 1.4 seconds to 0.2 seconds for 3D volumes of size (145, 174, 145) voxels with approximately 3000 keypoints. Notable speedups include the convolution operation (20X), 4D peak detection (3X), sub-sampling (3X), and difference-of-Gaussian pyramid construction (2X). Efficient descriptors offer a speedup of 2X and a memory savings of 6X compared to standard SIFT-Rank descriptors, at a cost of reduced numbers of keypoint correspondences, revealing a trade-off between computational efficiency and algorithmic performance. The speedups gained by our implementation will allow for a more efficient analysis on larger data sets. Our optimized GPU implementation of the 3D SIFT-Rank extractor is available at https://github.com/CarluerJB/3D_SIFT_CUDA.
Curating Subject ID Labels using Keypoint Signatures
Oct 07, 2021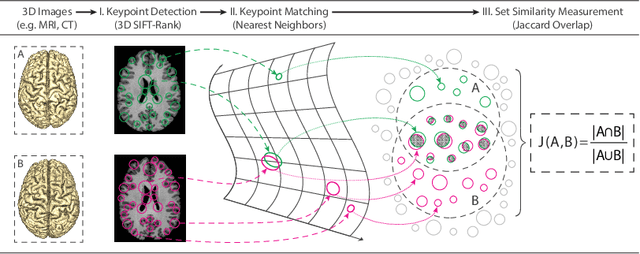

Abstract:Subject ID labels are unique, anonymized codes that can be used to group all images of a subject while maintaining anonymity. ID errors may be inadvertently introduced manually error during enrollment and may lead to systematic error into machine learning evaluation (e.g. due to double-dipping) or potential patient misdiagnosis in clinical contexts. Here we describe a highly efficient system for curating subject ID labels in large generic medical image datasets, based on the 3D image keypoint representation, which recently led to the discovery of previously unknown labeling errors in widely-used public brain MRI datasets
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge