William Wells III
Connecting Jensen-Shannon and Kullback-Leibler Divergences: A New Bound for Representation Learning
Oct 23, 2025Abstract:Mutual Information (MI) is a fundamental measure of statistical dependence widely used in representation learning. While direct optimization of MI via its definition as a Kullback-Leibler divergence (KLD) is often intractable, many recent methods have instead maximized alternative dependence measures, most notably, the Jensen-Shannon divergence (JSD) between joint and product of marginal distributions via discriminative losses. However, the connection between these surrogate objectives and MI remains poorly understood. In this work, we bridge this gap by deriving a new, tight, and tractable lower bound on KLD as a function of JSD in the general case. By specializing this bound to joint and marginal distributions, we demonstrate that maximizing the JSD-based information increases a guaranteed lower bound on mutual information. Furthermore, we revisit the practical implementation of JSD-based objectives and observe that minimizing the cross-entropy loss of a binary classifier trained to distinguish joint from marginal pairs recovers a known variational lower bound on the JSD. Extensive experiments demonstrate that our lower bound is tight when applied to MI estimation. We compared our lower bound to state-of-the-art neural estimators of variational lower bound across a range of established reference scenarios. Our lower bound estimator consistently provides a stable, low-variance estimate of a tight lower bound on MI. We also demonstrate its practical usefulness in the context of the Information Bottleneck framework. Taken together, our results provide new theoretical justifications and strong empirical evidence for using discriminative learning in MI-based representation learning.
Registering Image Volumes using 3D SIFT and Discrete SP-Symmetry
May 30, 2022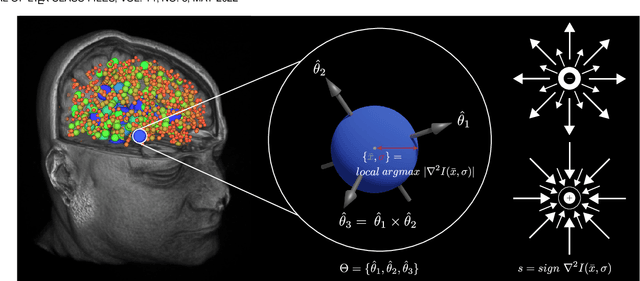
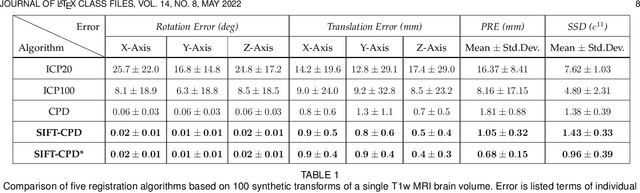
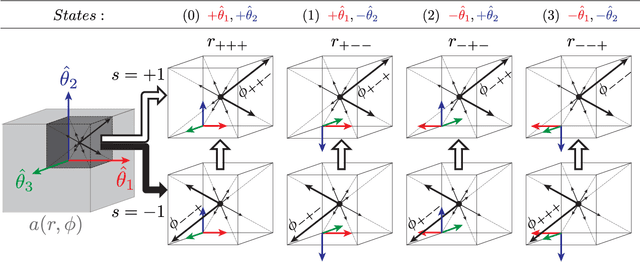
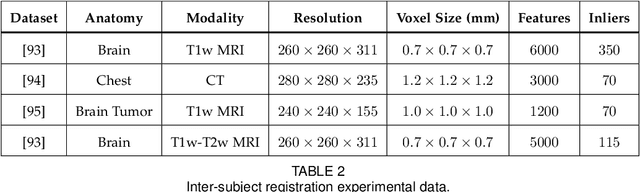
Abstract:This paper proposes to extend local image features in 3D to include invariance to discrete symmetry including inversion of spatial axes and image contrast. A binary feature sign $s \in \{-1,+1\}$ is defined as the sign of the Laplacian operator $\nabla^2$, and used to obtain a descriptor that is invariant to image sign inversion $s \rightarrow -s$ and 3D parity transforms $(x,y,z)\rightarrow(-x,-y,-z)$, i.e. SP-invariant or SP-symmetric. SP-symmetry applies to arbitrary scalar image fields $I: R^3 \rightarrow R^1$ mapping 3D coordinates $(x,y,z) \in R^3$ to scalar intensity $I(x,y,z) \in R^1$, generalizing the well-known charge conjugation and parity symmetry (CP-symmetry) applying to elementary charged particles. Feature orientation is modeled as a set of discrete states corresponding to potential axis reflections, independently of image contrast inversion. Two primary axis vectors are derived from image observations and potentially subject to reflection, and a third axis is an axial vector defined by the right-hand rule. Augmenting local feature properties with sign in addition to standard (location, scale, orientation) geometry leads to descriptors that are invariant to coordinate reflections and intensity contrast inversion. Feature properties are factored in to probabilistic point-based registration as symmetric kernels, based on a model of binary feature correspondence. Experiments using the well-known coherent point drift (CPD) algorithm demonstrate that SIFT-CPD kernels achieve the most accurate and rapid registration of the human brain and CT chest, including multiple MRI modalities of differing intensity contrast, and abnormal local variations such as tumors or occlusions. SIFT-CPD image registration is invariant to global scaling, rotation and translation and image intensity inversions of the input data.
Model and predict age and sex in healthy subjects using brain white matter features: A deep learning approach
Feb 08, 2022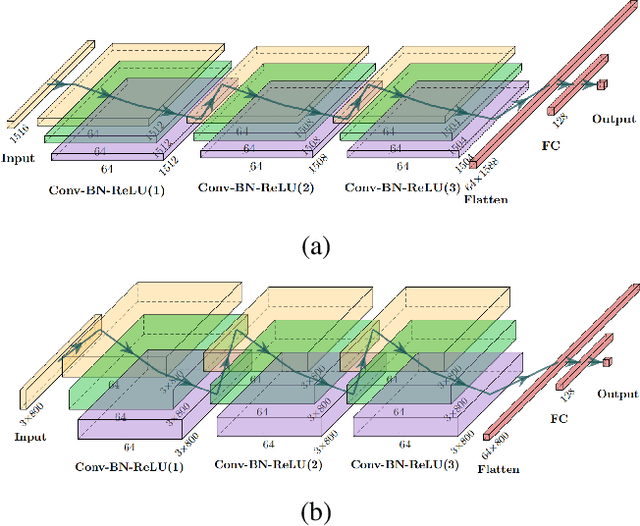
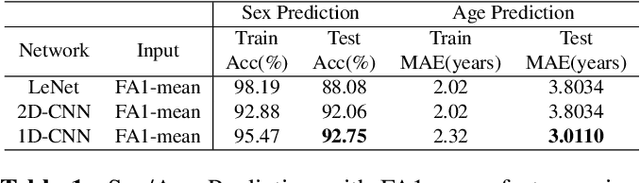
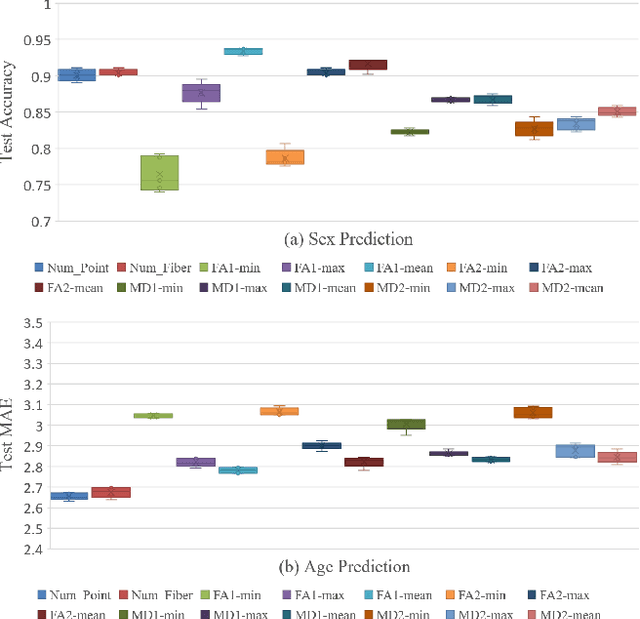
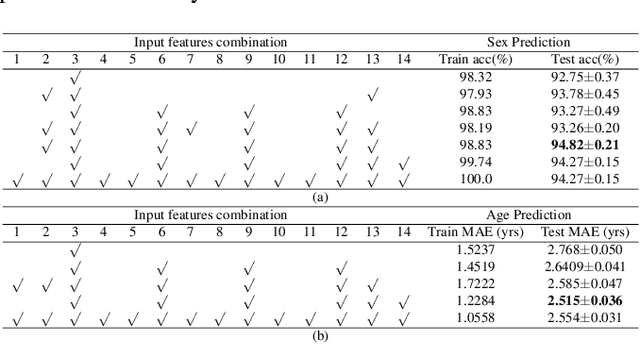
Abstract:The human brain's white matter (WM) structure is of immense interest to the scientific community. Diffusion MRI gives a powerful tool to describe the brain WM structure noninvasively. To potentially enable monitoring of age-related changes and investigation of sex-related brain structure differences on the mapping between the brain connectome and healthy subjects' age and sex, we extract fiber-cluster-based diffusion features and predict sex and age with a novel ensembled neural network classifier. We conduct experiments on the Human Connectome Project (HCP) young adult dataset and show that our model achieves 94.82% accuracy in sex prediction and 2.51 years MAE in age prediction. We also show that the fractional anisotropy (FA) is the most predictive of sex, while the number of fibers is the most predictive of age and the combination of different features can improve the model performance.
Double-Uncertainty Assisted Spatial and Temporal Regularization Weighting for Learning-based Registration
Jul 15, 2021


Abstract:In order to tackle the difficulty associated with the ill-posed nature of the image registration problem, researchers use regularization to constrain the solution space. For most learning-based registration approaches, the regularization usually has a fixed weight and only constrains the spatial transformation. Such convention has two limitations: (1) The regularization strength of a specific image pair should be associated with the content of the images, thus the ``one value fits all'' scheme is not ideal; (2) Only spatially regularizing the transformation (but overlooking the temporal consistency of different estimations) may not be the best strategy to cope with the ill-posedness. In this study, we propose a mean-teacher based registration framework. This framework incorporates an additional \textit{temporal regularization} term by encouraging the teacher model's temporal ensemble prediction to be consistent with that of the student model. At each training step, it also automatically adjusts the weights of the \textit{spatial regularization} and the \textit{temporal regularization} by taking account of the transformation uncertainty and appearance uncertainty derived from the perturbed teacher model. We perform experiments on multi- and uni-modal registration tasks, and the results show that our strategy outperforms the traditional and learning-based benchmark methods.
Efficient Pairwise Neuroimage Analysis using the Soft Jaccard Index and 3D Keypoint Sets
Mar 15, 2021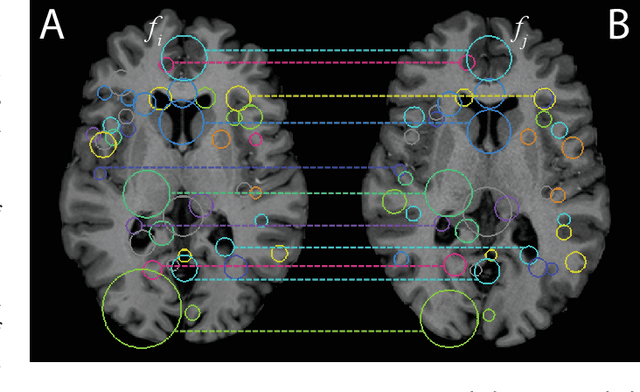
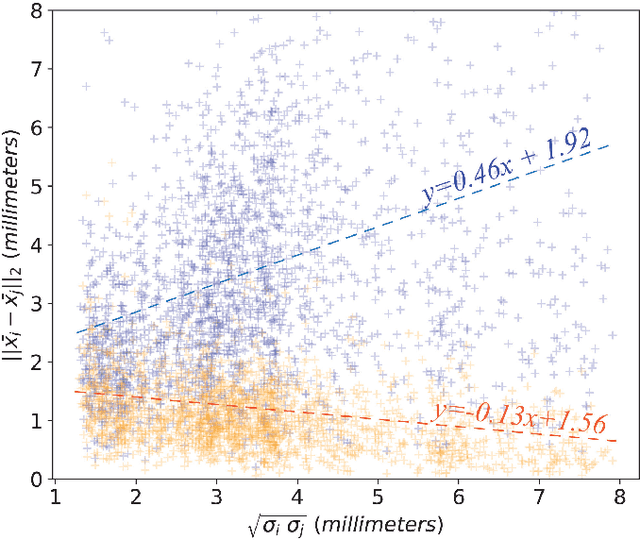
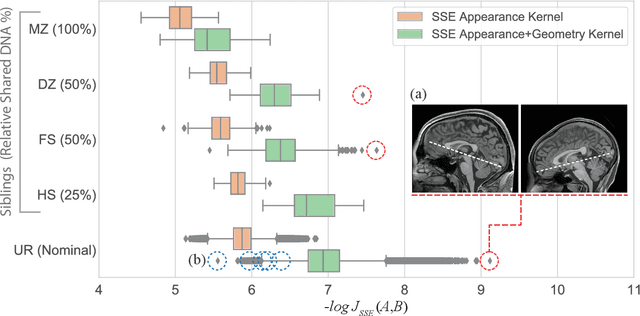
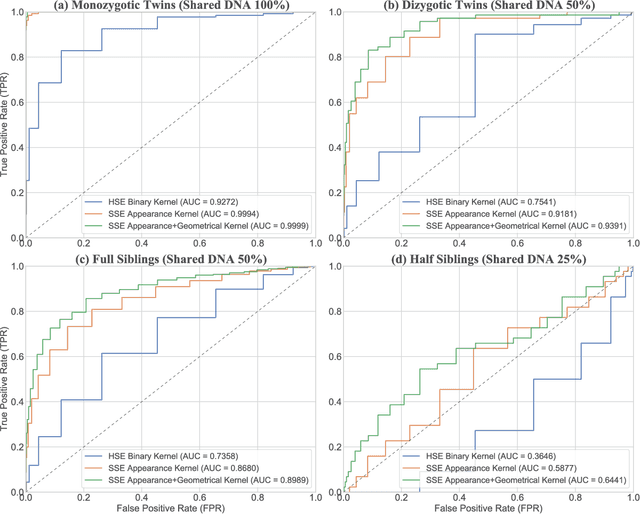
Abstract:We propose a novel pairwise distance measure between variable-sized sets of image keypoints for the purpose of large-scale medical image indexing. Our measure generalizes the Jaccard index to account for soft set equivalence (SSE) between set elements, via an adaptive kernel framework accounting for uncertainty in keypoint appearance and geometry. Novel kernels are proposed to quantify the variability of keypoint geometry in location and scale. Our distance measure may be estimated between $N^2$ image pairs in $O(N~\log~N)$ operations via keypoint indexing. Experiments validate our method in predicting 509,545 pairwise relationships from T1-weighted MRI brain volumes of monozygotic and dizygotic twins, siblings and half-siblings sharing 100%-25% of their polymorphic genes. Soft set equivalence and keypoint geometry kernels outperform standard hard set equivalence (HSE) in predicting family relationships. High accuracy is achieved, with monozygotic twin identification near 100% and several cases of unknown family labels, due to errors in the genotyping process, are correctly paired with family members. Software is provided for efficient fine-grained curation of large, generic image datasets.
Adversarial Uni- and Multi-modal Stream Networks for Multimodal Image Registration
Jul 06, 2020



Abstract:Deformable image registration between Computed Tomography (CT) images and Magnetic Resonance (MR) imaging is essential for many image-guided therapies. In this paper, we propose a novel translation-based unsupervised deformable image registration method. Distinct from other translation-based methods that attempt to convert the multimodal problem (e.g., CT-to-MR) into a unimodal problem (e.g., MR-to-MR) via image-to-image translation, our method leverages the deformation fields estimated from both: (i) the translated MR image and (ii) the original CT image in a dual-stream fashion, and automatically learns how to fuse them to achieve better registration performance. The multimodal registration network can be effectively trained by computationally efficient similarity metrics without any ground-truth deformation. Our method has been evaluated on two clinical datasets and demonstrates promising results compared to state-of-the-art traditional and learning-based methods.
Do Public Datasets Assure Unbiased Comparisons for Registration Evaluation?
Mar 20, 2020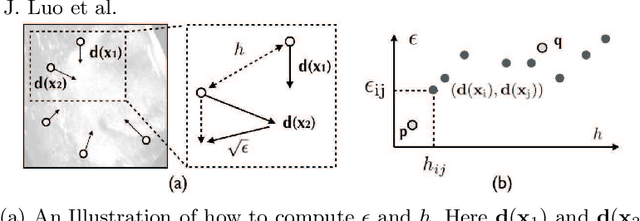
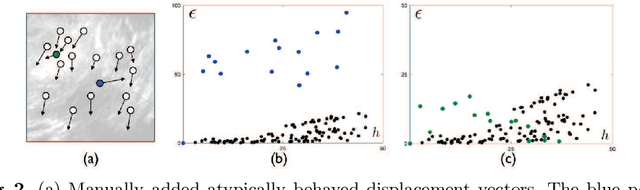
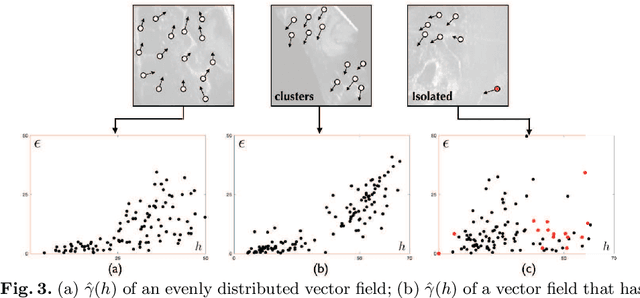
Abstract:With the increasing availability of new image registration approaches, an unbiased evaluation is becoming more needed so that clinicians can choose the most suitable approaches for their applications. Current evaluations typically use landmarks in manually annotated datasets. As a result, the quality of annotations is crucial for unbiased comparisons. Even though most data providers claim to have quality control over their datasets, an objective third-party screening can be reassuring for intended users. In this study, we use the variogram to screen the manually annotated landmarks in two datasets used to benchmark registration in image-guided neurosurgeries. The variogram provides an intuitive 2D representation of the spatial characteristics of annotated landmarks. Using variograms, we identified potentially problematic cases and had them examined by experienced radiologists. We found that (1) a small number of annotations may have fiducial localization errors; (2) the landmark distribution for some cases is not ideal to offer fair comparisons. If unresolved, both findings could incur bias in registration evaluation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge