Laurent Chauvin
Using Atom-Like Local Image Features to Study Human Genetics and Neuroanatomy in Large Sets of 3D Medical Image Volumes
Aug 25, 2022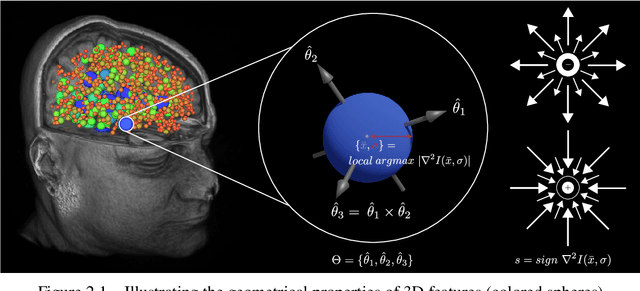
Abstract:The contributions of this thesis stem from technology developed to analyse large sets of volumetric images in terms of atom-like features extracted in 3D image space, following SIFT algorithm in 2D image space. New feature properties are introduced including a binary feature sign, analogous to an electrical charge, and a discrete set of symmetric feature orientation states in 3D space. These new properties are leveraged to extend feature invariance to include the sign inversion and parity (SP) transform, analogous to the charge conjugation and parity (CP) transform between a particle and its antiparticle in quantum mechanics, thereby accounting for local intensity contrast inversion between imaging modalities and axis reflections due to shape symmetry. A novel exponential kernel is proposed to quantify the similarity of a pair of features extracted in different images from their properties including location, scale, orientation, sign and appearance. A novel measure entitled the soft Jaccard is proposed to quantify the similarity of a pair of feature sets based on their overlap or intersection-over-union, where a kernel establishes non-binary or soft equivalence between a pair of feature elements. The soft Jaccard may be used to identify pairs of feature sets extracted from the same individuals or families with high accuracy, and a simple distance threshold led to the surprising discovery of previously unknown individual and family labeling errors in major public neuroimage datasets. A new algorithm is proposed to register or spatially align a pair of feature sets, entitled SIFT Coherent Point Drift (SIFT-CPD), by identifying a transform that maximizes the soft Jaccard between a fixed feature set and a transformed set. SIFT-CPD achieves faster and more accurate registration than the original CPD algorithm based on feature location information alone, in a variety of challenging.
Registering Image Volumes using 3D SIFT and Discrete SP-Symmetry
May 30, 2022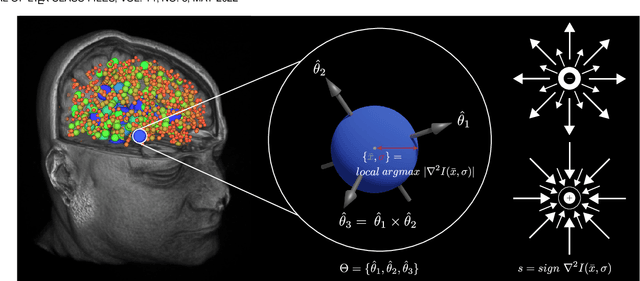
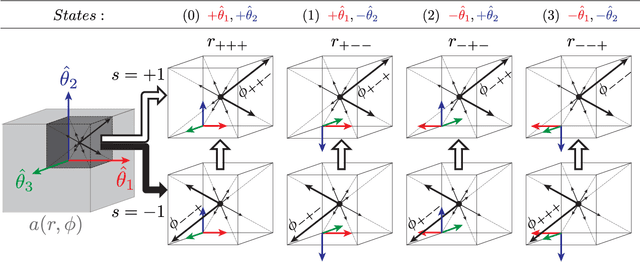
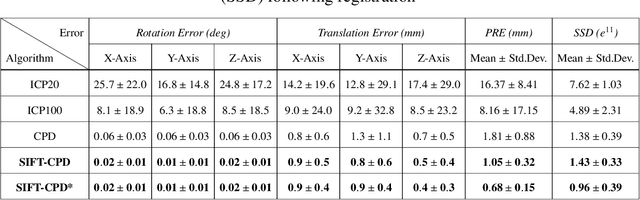
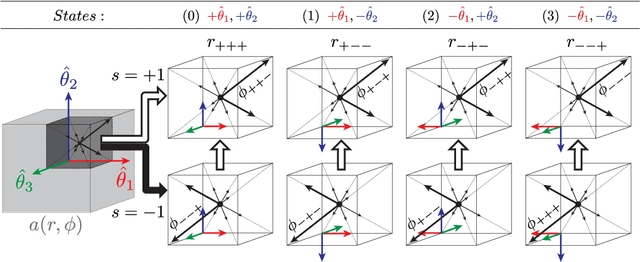
Abstract:This paper proposes to extend local image features in 3D to include invariance to discrete symmetry including inversion of spatial axes and image contrast. A binary feature sign $s \in \{-1,+1\}$ is defined as the sign of the Laplacian operator $\nabla^2$, and used to obtain a descriptor that is invariant to image sign inversion $s \rightarrow -s$ and 3D parity transforms $(x,y,z)\rightarrow(-x,-y,-z)$, i.e. SP-invariant or SP-symmetric. SP-symmetry applies to arbitrary scalar image fields $I: R^3 \rightarrow R^1$ mapping 3D coordinates $(x,y,z) \in R^3$ to scalar intensity $I(x,y,z) \in R^1$, generalizing the well-known charge conjugation and parity symmetry (CP-symmetry) applying to elementary charged particles. Feature orientation is modeled as a set of discrete states corresponding to potential axis reflections, independently of image contrast inversion. Two primary axis vectors are derived from image observations and potentially subject to reflection, and a third axis is an axial vector defined by the right-hand rule. Augmenting local feature properties with sign in addition to standard (location, scale, orientation) geometry leads to descriptors that are invariant to coordinate reflections and intensity contrast inversion. Feature properties are factored in to probabilistic point-based registration as symmetric kernels, based on a model of binary feature correspondence. Experiments using the well-known coherent point drift (CPD) algorithm demonstrate that SIFT-CPD kernels achieve the most accurate and rapid registration of the human brain and CT chest, including multiple MRI modalities of differing intensity contrast, and abnormal local variations such as tumors or occlusions. SIFT-CPD image registration is invariant to global scaling, rotation and translation and image intensity inversions of the input data.
GPU optimization of the 3D Scale-invariant Feature Transform Algorithm and a Novel BRIEF-inspired 3D Fast Descriptor
Dec 19, 2021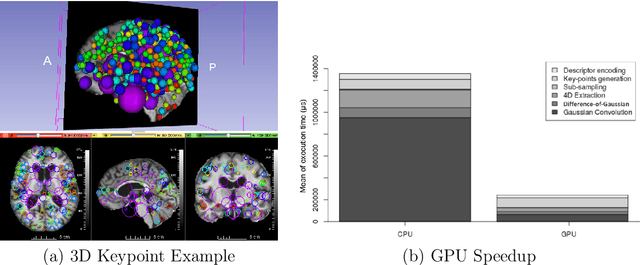
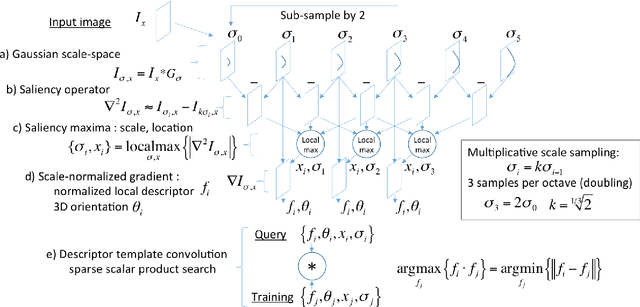
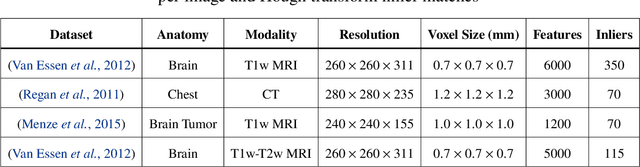
Abstract:This work details a highly efficient implementation of the 3D scale-invariant feature transform (SIFT) algorithm, for the purpose of machine learning from large sets of volumetric medical image data. The primary operations of the 3D SIFT code are implemented on a graphics processing unit (GPU), including convolution, sub-sampling, and 4D peak detection from scale-space pyramids. The performance improvements are quantified in keypoint detection and image-to-image matching experiments, using 3D MRI human brain volumes of different people. Computationally efficient 3D keypoint descriptors are proposed based on the Binary Robust Independent Elementary Feature (BRIEF) code, including a novel descriptor we call Ranked Robust Independent Elementary Features (RRIEF), and compared to the original 3D SIFT-Rank method\citep{toews2013efficient}. The GPU implementation affords a speedup of approximately 7X beyond an optimised CPU implementation, where computation time is reduced from 1.4 seconds to 0.2 seconds for 3D volumes of size (145, 174, 145) voxels with approximately 3000 keypoints. Notable speedups include the convolution operation (20X), 4D peak detection (3X), sub-sampling (3X), and difference-of-Gaussian pyramid construction (2X). Efficient descriptors offer a speedup of 2X and a memory savings of 6X compared to standard SIFT-Rank descriptors, at a cost of reduced numbers of keypoint correspondences, revealing a trade-off between computational efficiency and algorithmic performance. The speedups gained by our implementation will allow for a more efficient analysis on larger data sets. Our optimized GPU implementation of the 3D SIFT-Rank extractor is available at https://github.com/CarluerJB/3D_SIFT_CUDA.
Curating Subject ID Labels using Keypoint Signatures
Oct 07, 2021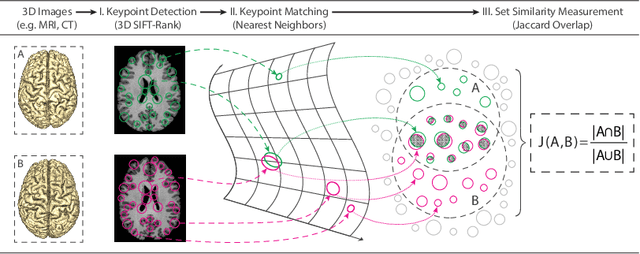

Abstract:Subject ID labels are unique, anonymized codes that can be used to group all images of a subject while maintaining anonymity. ID errors may be inadvertently introduced manually error during enrollment and may lead to systematic error into machine learning evaluation (e.g. due to double-dipping) or potential patient misdiagnosis in clinical contexts. Here we describe a highly efficient system for curating subject ID labels in large generic medical image datasets, based on the 3D image keypoint representation, which recently led to the discovery of previously unknown labeling errors in widely-used public brain MRI datasets
Efficient Pairwise Neuroimage Analysis using the Soft Jaccard Index and 3D Keypoint Sets
Mar 15, 2021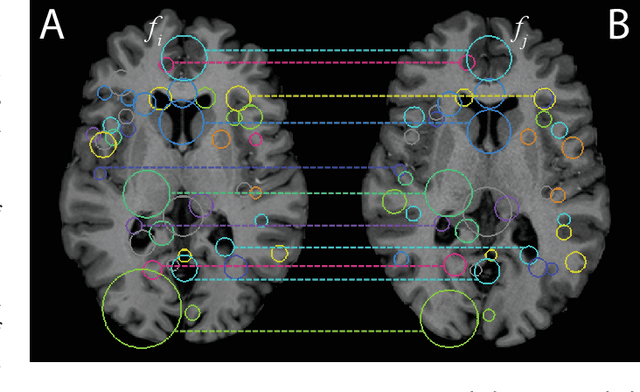
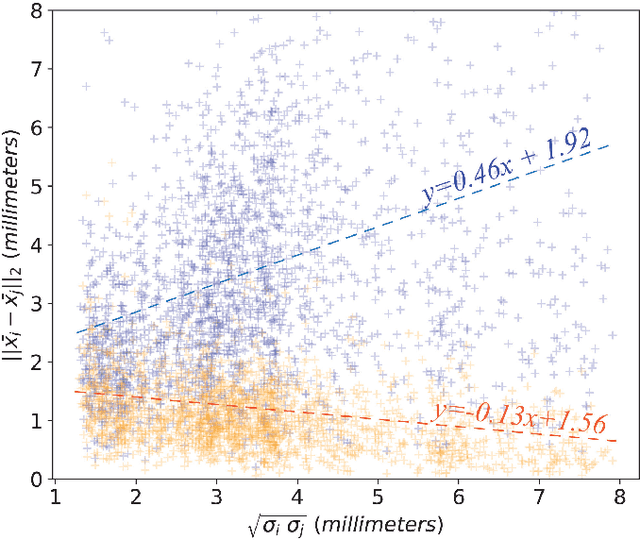
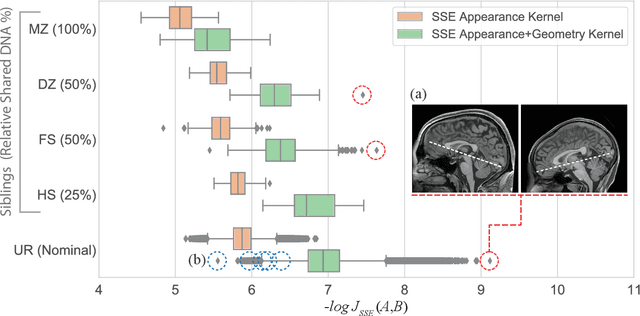
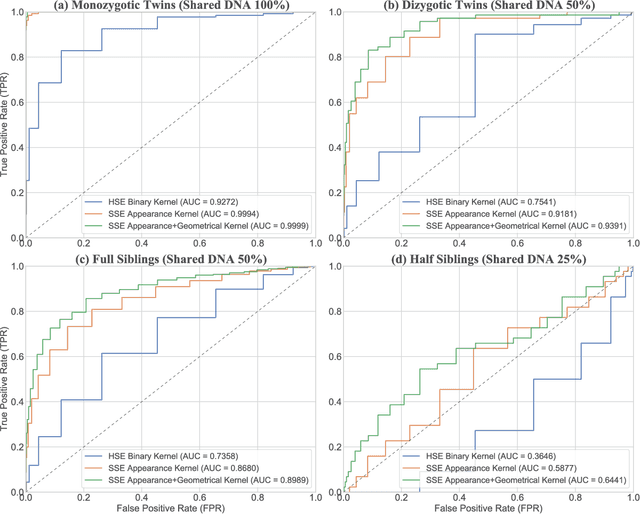
Abstract:We propose a novel pairwise distance measure between variable-sized sets of image keypoints for the purpose of large-scale medical image indexing. Our measure generalizes the Jaccard index to account for soft set equivalence (SSE) between set elements, via an adaptive kernel framework accounting for uncertainty in keypoint appearance and geometry. Novel kernels are proposed to quantify the variability of keypoint geometry in location and scale. Our distance measure may be estimated between $N^2$ image pairs in $O(N~\log~N)$ operations via keypoint indexing. Experiments validate our method in predicting 509,545 pairwise relationships from T1-weighted MRI brain volumes of monozygotic and dizygotic twins, siblings and half-siblings sharing 100%-25% of their polymorphic genes. Soft set equivalence and keypoint geometry kernels outperform standard hard set equivalence (HSE) in predicting family relationships. High accuracy is achieved, with monozygotic twin identification near 100% and several cases of unknown family labels, due to errors in the genotyping process, are correctly paired with family members. Software is provided for efficient fine-grained curation of large, generic image datasets.
Multi-modal analysis of genetically-related subjects using SIFT descriptors in brain MRI
Sep 18, 2017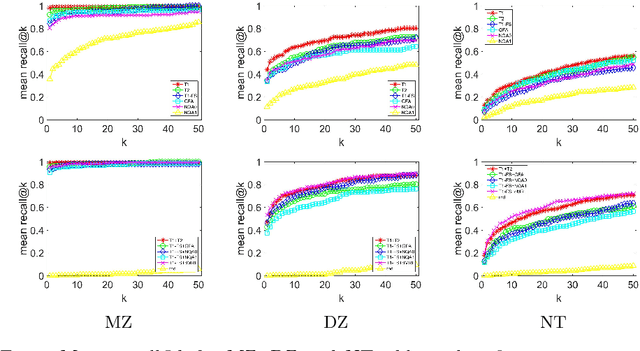
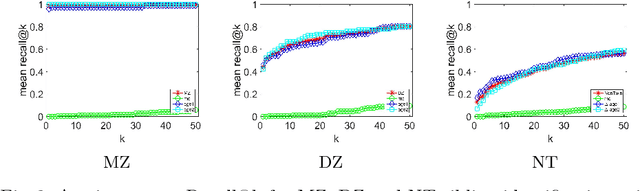
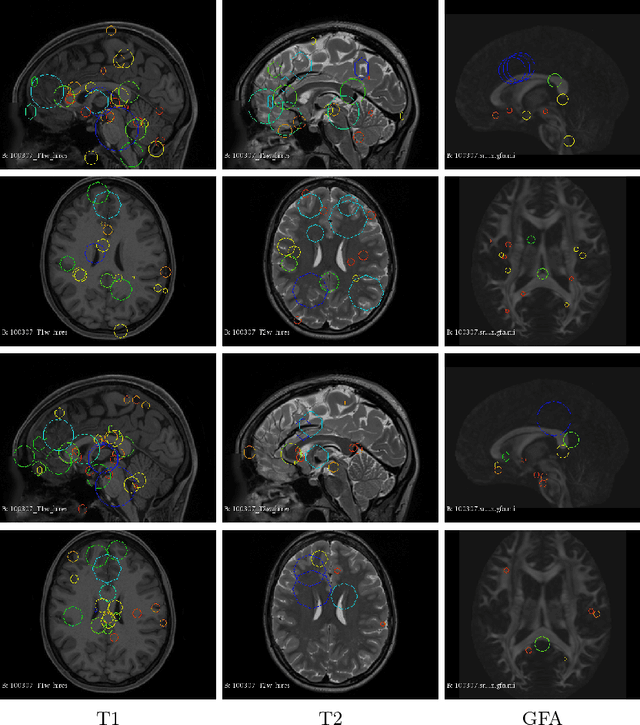
Abstract:So far, fingerprinting studies have focused on identifying features from single-modality MRI data, which capture individual characteristics in terms of brain structure, function, or white matter microstructure. However, due to the lack of a framework for comparing across multiple modalities, studies based on multi-modal data remain elusive. This paper presents a multi-modal analysis of genetically-related subjects to compare and contrast the information provided by various MRI modalities. The proposed framework represents MRI scans as bags of SIFT features, and uses these features in a nearest-neighbor graph to measure subject similarity. Experiments using the T1/T2-weighted MRI and diffusion MRI data of 861 Human Connectome Project subjects demonstrate strong links between the proposed similarity measure and genetic proximity.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge