Kuldeep Kumar
Efficient Pairwise Neuroimage Analysis using the Soft Jaccard Index and 3D Keypoint Sets
Mar 15, 2021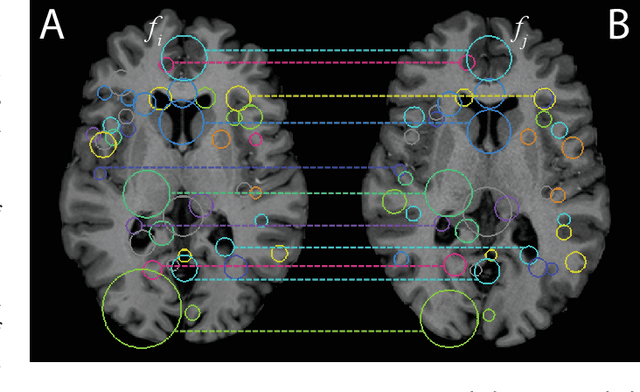
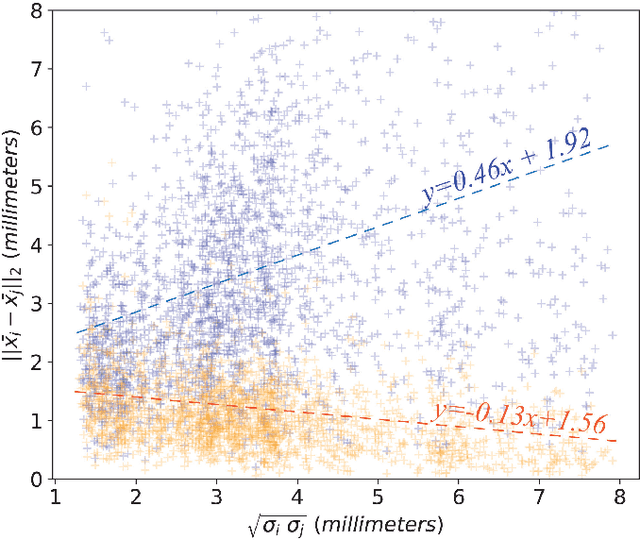
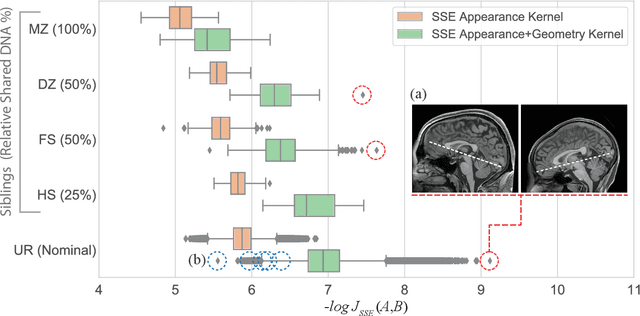
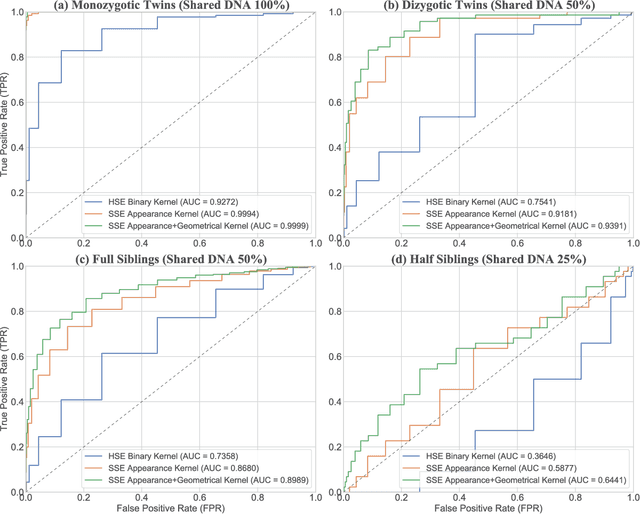
Abstract:We propose a novel pairwise distance measure between variable-sized sets of image keypoints for the purpose of large-scale medical image indexing. Our measure generalizes the Jaccard index to account for soft set equivalence (SSE) between set elements, via an adaptive kernel framework accounting for uncertainty in keypoint appearance and geometry. Novel kernels are proposed to quantify the variability of keypoint geometry in location and scale. Our distance measure may be estimated between $N^2$ image pairs in $O(N~\log~N)$ operations via keypoint indexing. Experiments validate our method in predicting 509,545 pairwise relationships from T1-weighted MRI brain volumes of monozygotic and dizygotic twins, siblings and half-siblings sharing 100%-25% of their polymorphic genes. Soft set equivalence and keypoint geometry kernels outperform standard hard set equivalence (HSE) in predicting family relationships. High accuracy is achieved, with monozygotic twin identification near 100% and several cases of unknown family labels, due to errors in the genotyping process, are correctly paired with family members. Software is provided for efficient fine-grained curation of large, generic image datasets.
White matter fiber analysis using kernel dictionary learning and sparsity priors
Apr 15, 2018



Abstract:Diffusion magnetic resonance imaging, a non-invasive tool to infer white matter fiber connections, produces a large number of streamlines containing a wealth of information on structural connectivity. The size of these tractography outputs makes further analyses complex, creating a need for methods to group streamlines into meaningful bundles. In this work, we address this by proposing a set of kernel dictionary learning and sparsity priors based methods. Proposed frameworks include L-0 norm, group sparsity, as well as manifold regularization prior. The proposed methods allow streamlines to be assigned to more than one bundle, making it more robust to overlapping bundles and inter-subject variations. We evaluate the performance of our method on a labeled set and data from Human Connectome Project. Results highlight the ability of our method to group streamlines into plausible bundles and illustrate the impact of sparsity priors on the performance of the proposed methods.
Multi-modal analysis of genetically-related subjects using SIFT descriptors in brain MRI
Sep 18, 2017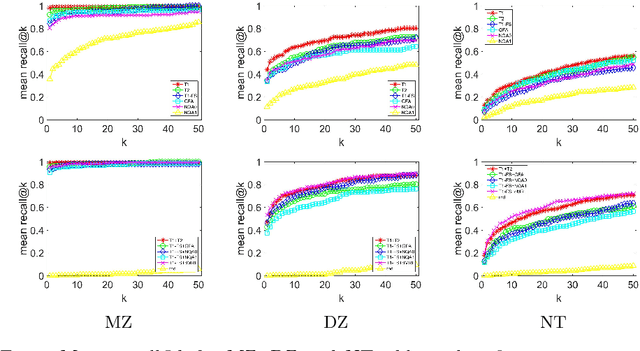
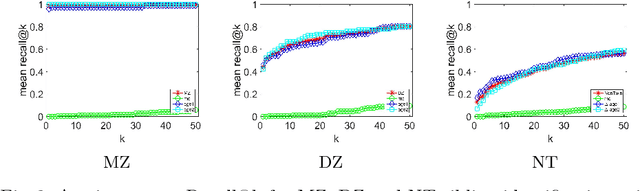
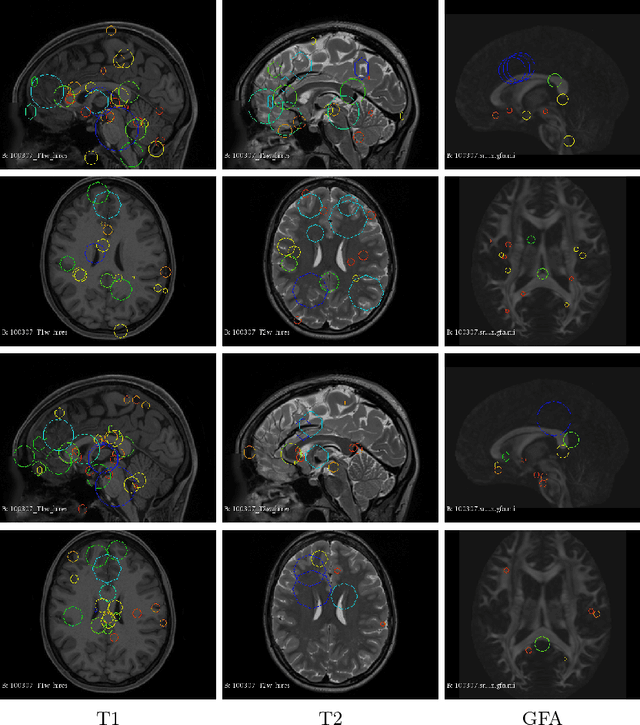
Abstract:So far, fingerprinting studies have focused on identifying features from single-modality MRI data, which capture individual characteristics in terms of brain structure, function, or white matter microstructure. However, due to the lack of a framework for comparing across multiple modalities, studies based on multi-modal data remain elusive. This paper presents a multi-modal analysis of genetically-related subjects to compare and contrast the information provided by various MRI modalities. The proposed framework represents MRI scans as bags of SIFT features, and uses these features in a nearest-neighbor graph to measure subject similarity. Experiments using the T1/T2-weighted MRI and diffusion MRI data of 861 Human Connectome Project subjects demonstrate strong links between the proposed similarity measure and genetic proximity.
White Matter Fiber Segmentation Using Functional Varifolds
Sep 18, 2017


Abstract:The extraction of fibers from dMRI data typically produces a large number of fibers, it is common to group fibers into bundles. To this end, many specialized distance measures, such as MCP, have been used for fiber similarity. However, these distance based approaches require point-wise correspondence and focus only on the geometry of the fibers. Recent publications have highlighted that using microstructure measures along fibers improves tractography analysis. Also, many neurodegenerative diseases impacting white matter require the study of microstructure measures as well as the white matter geometry. Motivated by these, we propose to use a novel computational model for fibers, called functional varifolds, characterized by a metric that considers both the geometry and microstructure measure (e.g. GFA) along the fiber pathway. We use it to cluster fibers with a dictionary learning and sparse coding-based framework, and present a preliminary analysis using HCP data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge