Stanley Durrleman
ARAMIS
Benchmarking off-the-shelf statistical shape modeling tools in clinical applications
Sep 07, 2020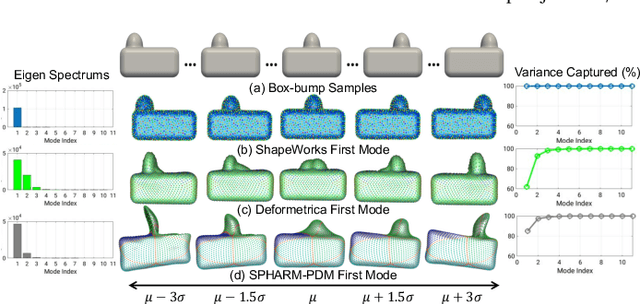

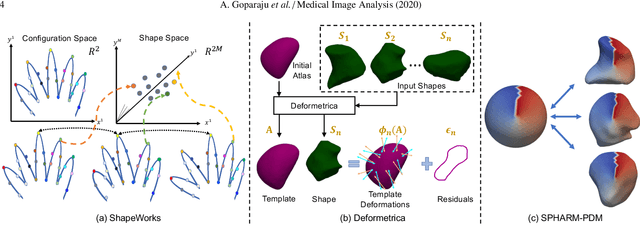
Abstract:Statistical shape modeling (SSM) is widely used in biology and medicine as a new generation of morphometric approaches for the quantitative analysis of anatomical shapes. Technological advancements of in vivo imaging have led to the development of open-source computational tools that automate the modeling of anatomical shapes and their population-level variability. However, little work has been done on the evaluation and validation of such tools in clinical applications that rely on morphometric quantifications (e.g., implant design and lesion screening). Here, we systematically assess the outcome of widely used, state-of-the-art SSM tools, namely ShapeWorks, Deformetrica, and SPHARM-PDM. We use both quantitative and qualitative metrics to evaluate shape models from different tools. We propose validation frameworks for anatomical landmark/measurement inference and lesion screening. We also present a lesion screening method to objectively characterize subtle abnormal shape changes with respect to learned population-level statistics of controls. Results demonstrate that SSM tools display different levels of consistencies, where ShapeWorks and Deformetrica models are more consistent compared to models from SPHARM-PDM due to the groupwise approach of estimating surface correspondences. Furthermore, ShapeWorks and Deformetrica shape models are found to capture clinically relevant population-level variability compared to SPHARM-PDM models.
Mixture of Conditional Gaussian Graphical Models for unlabelled heterogeneous populations in the presence of co-factors
Jun 19, 2020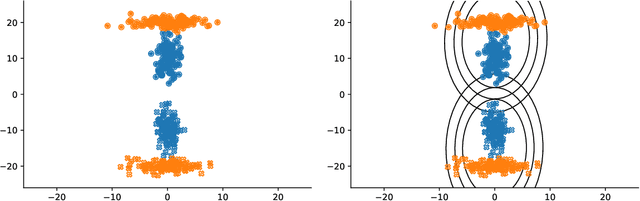
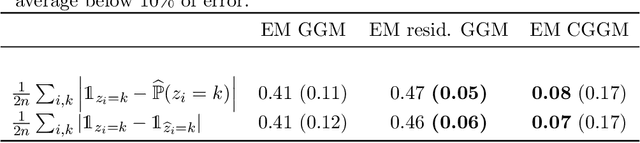
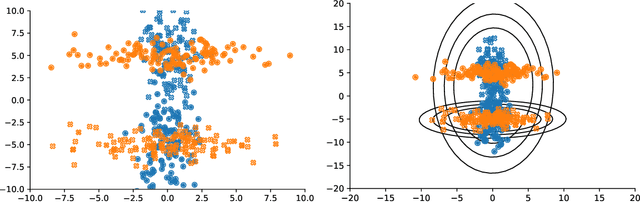
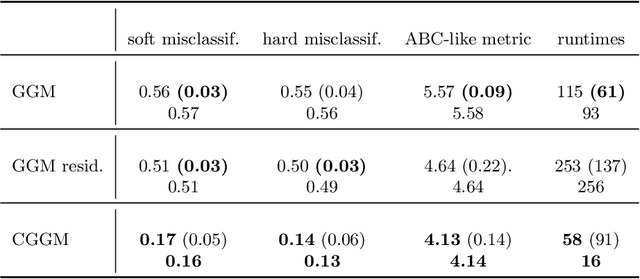
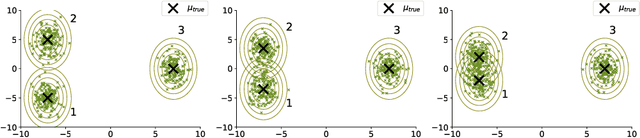
Abstract:Conditional correlation networks, within Gaussian Graphical Models (GGM), are widely used to describe the direct interactions between the components of a random vector. In the case of an unlabelled Heterogeneous population, Expectation Maximisation (EM) algorithms for Mixtures of GGM have been proposed to estimate both each sub-population's graph and the class labels. However, we argue that, with most real data, class affiliation cannot be described with a Mixture of Gaussian, which mostly groups data points according to their geometrical proximity. In particular, there often exists external co-features whose values affect the features' average value, scattering across the feature space data points belonging to the same sub-population. Additionally, if the co-features' effect on the features is Heterogeneous, then the estimation of this effect cannot be separated from the sub-population identification. In this article, we propose a Mixture of Conditional GGM (CGGM) that subtracts the heterogeneous effects of the co-features to regroup the data points into sub-population corresponding clusters. We develop a penalised EM algorithm to estimate graph-sparse model parameters. We demonstrate on synthetic and real data how this method fulfils its goal and succeeds in identifying the sub-populations where the Mixtures of GGM are disrupted by the effect of the co-features.
Deterministic Approximate EM Algorithm; Application to the Riemann Approximation EM and the Tempered EM
Mar 23, 2020
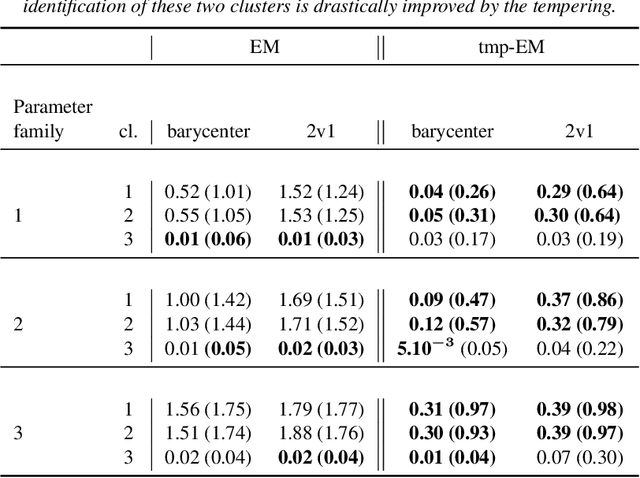
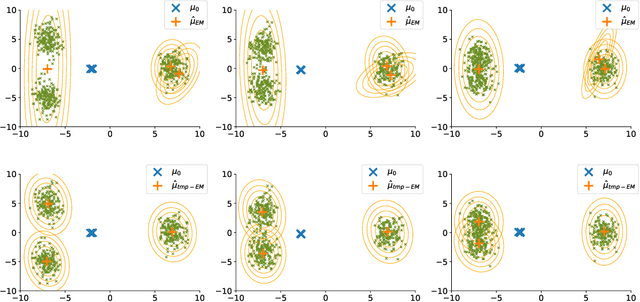
Abstract:The Expectation Maximisation (EM) algorithm is widely used to optimise non-convex likelihood functions with hidden variables. Many authors modified its simple design to fit more specific situations. For instance the Expectation (E) step has been replaced by Monte Carlo (MC) approximations, Markov Chain Monte Carlo approximations, tempered approximations... Most of the well studied approximations belong to the stochastic class. By comparison, the literature is lacking when it comes to deterministic approximations. In this paper, we introduce a theoretical framework, with state of the art convergence guarantees, for any deterministic approximation of the E step. We analyse theoretically and empirically several approximations that fit into this framework. First, for cases with intractable E steps, we introduce a deterministic alternative to the MC-EM, using Riemann sums. This method is easy to implement and does not require the tuning of hyper-parameters. Then, we consider the tempered approximation, borrowed from the Simulated Annealing optimisation technique and meant to improve the EM solution. We prove that the the tempered EM verifies the convergence guarantees for a wide range of temperature profiles. We showcase empirically how it is able to escape adversarial initialisations. Finally, we combine the Riemann and tempered approximations to accomplish both their purposes.
Gaussian Graphical Model exploration and selection in high dimension low sample size setting
Mar 11, 2020
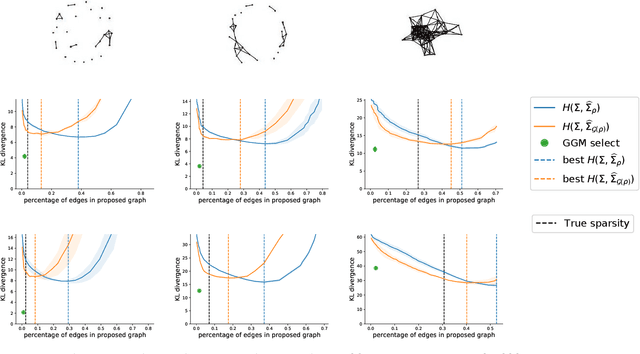
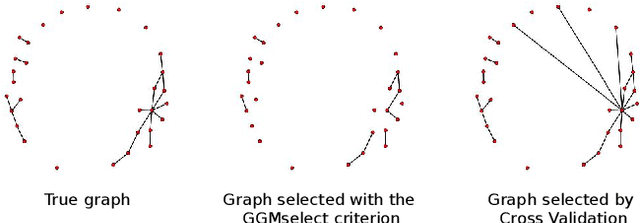
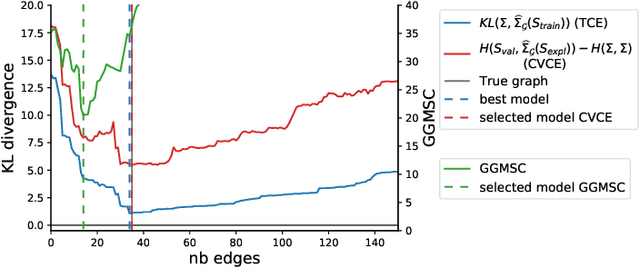
Abstract:Gaussian Graphical Models (GGM) are often used to describe the conditional correlations between the components of a random vector. In this article, we compare two families of GGM inference methods: nodewise edge selection and penalised likelihood maximisation. We demonstrate on synthetic data that, when the sample size is small, the two methods produce graphs with either too few or too many edges when compared to the real one. As a result, we propose a composite procedure that explores a family of graphs with an nodewise numerical scheme and selects a candidate among them with an overall likelihood criterion. We demonstrate that, when the number of observations is small, this selection method yields graphs closer to the truth and corresponding to distributions with better KL divergence with regards to the real distribution than the other two. Finally, we show the interest of our algorithm on two concrete cases: first on brain imaging data, then on biological nephrology data. In both cases our results are more in line with current knowledge in each field.
Convolutional Neural Networks for Classification of Alzheimer's Disease: Overview and Reproducible Evaluation
Apr 16, 2019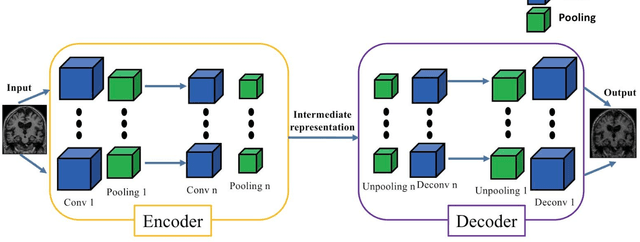
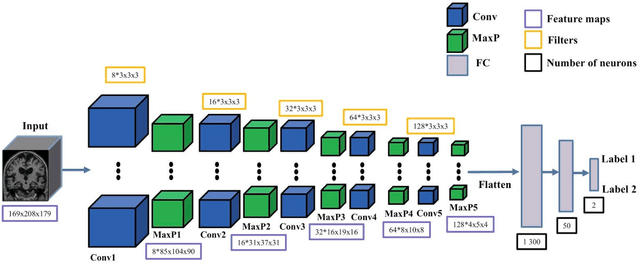
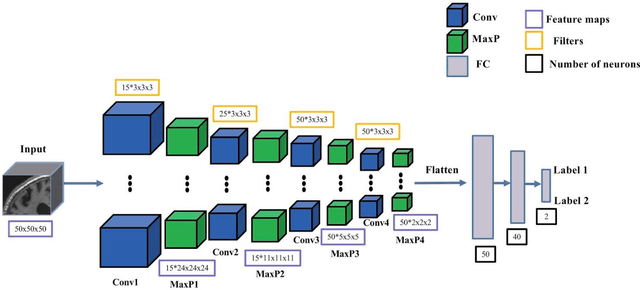
Abstract:In the past two years, over 30 papers have proposed to use convolutional neural network (CNN) for AD classification. However, the classification performances across studies are difficult to compare. Moreover, these studies are hardly reproducible because their frameworks are not publicly accessible. Lastly, some of these papers may reported biased performances due to inadequate or unclear validation procedure and also it is unclear how the model architecture and parameters were chosen. In the present work, we aim to address these limitations through three main contributions. First, we performed a systematic literature review of studies using CNN for AD classification from anatomical MRI. We identified four main types of approaches: 2D slice-level, 3D patch-level, ROI-based and 3D subject-level CNN. Moreover, we found that more than half of the surveyed papers may have suffered from data leakage and thus reported biased performances. Our second contribution is an open-source framework for classification of AD. Thirdly, we used this framework to rigorously compare different CNN architectures, which are representative of the existing literature, and to study the influence of key components on classification performances. On the validation set, the ROI-based (hippocampus) CNN achieved highest balanced accuracy (0.86 for AD vs CN and 0.80 for sMCI vs pMCI) compared to other approaches. Transfer learning with autoencoder pre-training did not improve the average accuracy but reduced the variance. Training using longitudinal data resulted in similar or higher performance, depending on the approach, compared to training with only baseline data. Sophisticated image preprocessing did not improve the results. Lastly, CNN performed similarly to standard SVM for task AD vs CN but outperformed SVM for task sMCI vs pMCI, demonstrating the potential of deep learning for challenging diagnostic tasks.
Simulation of virtual cohorts increases predictive accuracy of cognitive decline in MCI subjects
Apr 05, 2019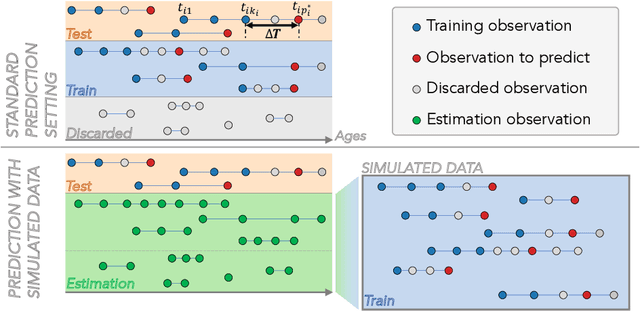
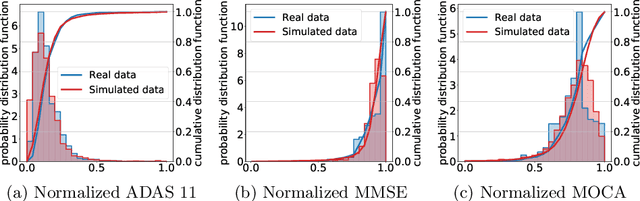
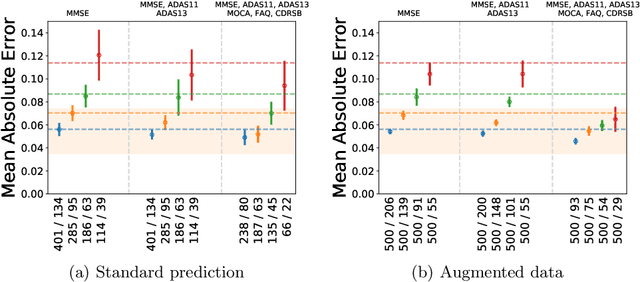
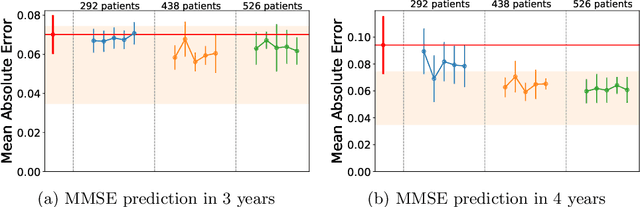
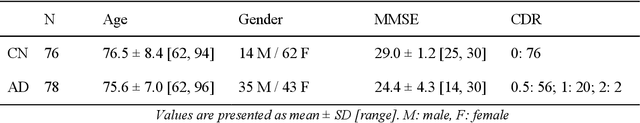
Abstract:The ability to predict the progression of biomarkers, notably in NDD, is limited by the size of the longitudinal data sets, in terms of number of patients, number of visits per patients and total follow-up time. To this end, we introduce a data augmentation technique that is able to reproduce the variability seen in a longitudinal training data set and simulate continuous biomarkers trajectories for any number of virtual patients. Thanks to this simulation framework, we propose to transform the training set into a simulated data set with more patients, more time-points per patient and longer follow-up duration. We illustrate this approach on the prediction of the MMSE of MCI subjects of the ADNI data set. We show that it allows to reach predictions with errors comparable to the noise in the data, estimated in test/retest studies, achieving a improvement of 37% of the mean absolute error compared to the same non-augmented model.
Reproducible evaluation of diffusion MRI features for automatic classification of patients with Alzheimers disease
Dec 28, 2018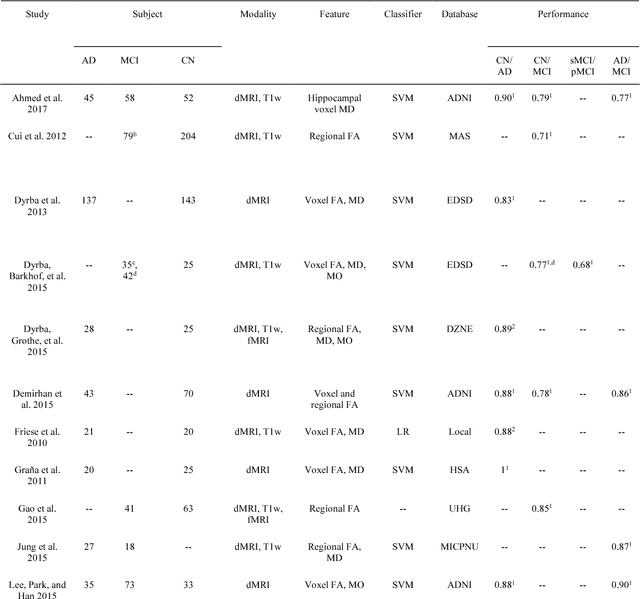
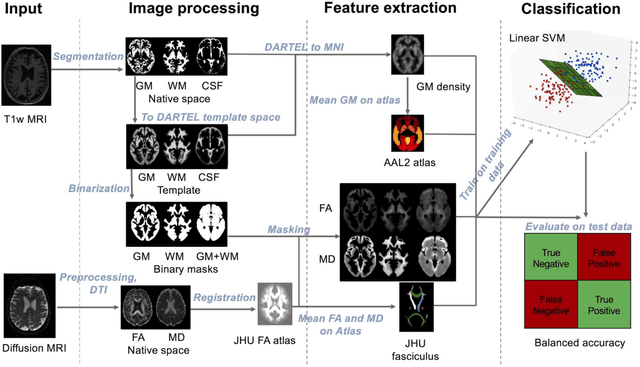
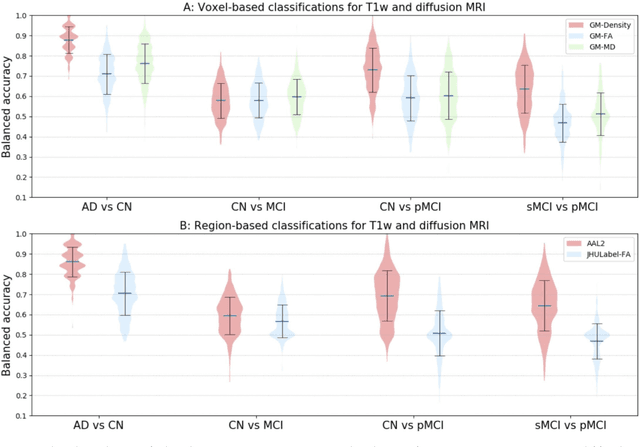
Abstract:Diffusion MRI is the modality of choice to study alterations of white matter. In the past years, various works have used diffusion MRI for automatic classification of Alzheimers disease. However, the performances obtained with different approaches are difficult to compare because of variations in components such as input data, participant selection, image preprocessing, feature extraction, feature selection (FS) and cross-validation (CV) procedure. Moreover, these studies are also difficult to reproduce because these different components are not readily available. In a previous work (Samper-Gonzalez et al. 2018), we proposed an open-source framework for the reproducible evaluation of AD classification from T1-weighted (T1w) MRI and PET data. In the present paper, we extend this framework to diffusion MRI data. The framework comprises: tools to automatically convert ADNI data into the BIDS standard, pipelines for image preprocessing and feature extraction, baseline classifiers and a rigorous CV procedure. We demonstrate the use of the framework through assessing the influence of diffusion tensor imaging (DTI) metrics (fractional anisotropy - FA, mean diffusivity - MD), feature types, imaging modalities (diffusion MRI or T1w MRI), data imbalance and FS bias. First, voxel-wise features generally gave better performances than regional features. Secondly, FA and MD provided comparable results for voxel-wise features. Thirdly, T1w MRI performed better than diffusion MRI. Fourthly, we demonstrated that using non-nested validation of FS leads to unreliable and over-optimistic results. All the code is publicly available: general-purpose tools have been integrated into the Clinica software (www.clinica.run) and the paper-specific code is available at: https://gitlab.icm-institute.org/aramislab/AD-ML.
Reproducible evaluation of classification methods in Alzheimer's disease: framework and application to MRI and PET data
Aug 20, 2018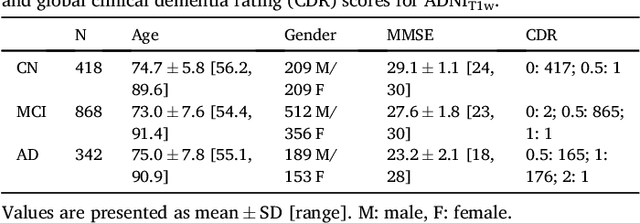
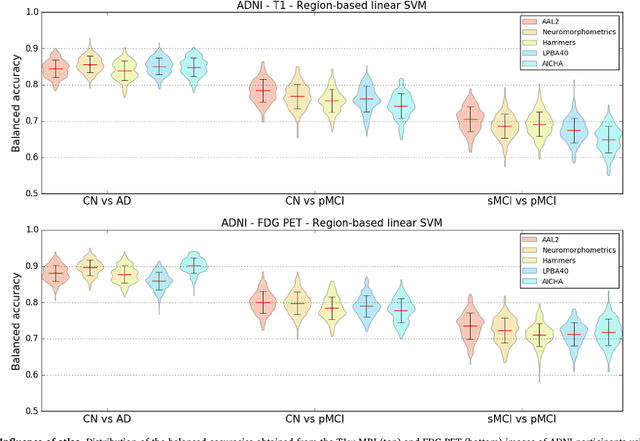
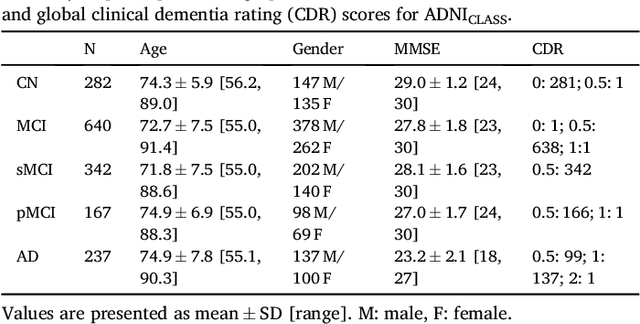
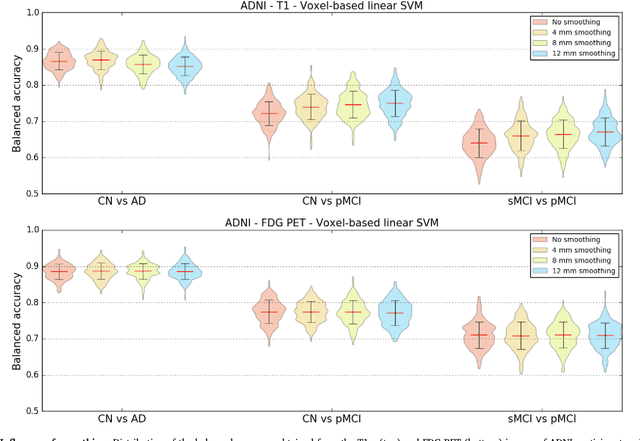
Abstract:A large number of papers have introduced novel machine learning and feature extraction methods for automatic classification of AD. However, they are difficult to reproduce because key components of the validation are often not readily available. These components include selected participants and input data, image preprocessing and cross-validation procedures. The performance of the different approaches is also difficult to compare objectively. In particular, it is often difficult to assess which part of the method provides a real improvement, if any. We propose a framework for reproducible and objective classification experiments in AD using three publicly available datasets (ADNI, AIBL and OASIS). The framework comprises: i) automatic conversion of the three datasets into BIDS format, ii) a modular set of preprocessing pipelines, feature extraction and classification methods, together with an evaluation framework, that provide a baseline for benchmarking the different components. We demonstrate the use of the framework for a large-scale evaluation on 1960 participants using T1 MRI and FDG PET data. In this evaluation, we assess the influence of different modalities, preprocessing, feature types, classifiers, training set sizes and datasets. Performances were in line with the state-of-the-art. FDG PET outperformed T1 MRI for all classification tasks. No difference in performance was found for the use of different atlases, image smoothing, partial volume correction of FDG PET images, or feature type. Linear SVM and L2-logistic regression resulted in similar performance and both outperformed random forests. The classification performance increased along with the number of subjects used for training. Classifiers trained on ADNI generalized well to AIBL and OASIS. All the code of the framework and the experiments is publicly available at: https://gitlab.icm-institute.org/aramislab/AD-ML.
Learning distributions of shape trajectories from longitudinal datasets: a hierarchical model on a manifold of diffeomorphisms
Jun 13, 2018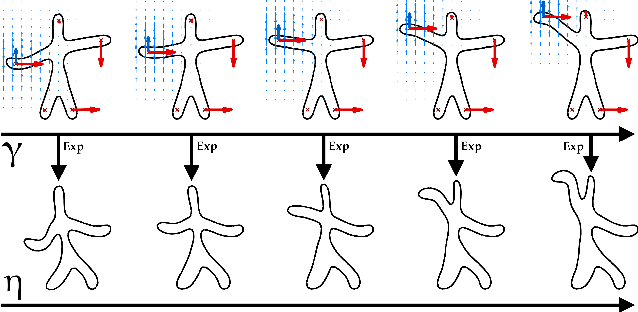


Abstract:We propose a method to learn a distribution of shape trajectories from longitudinal data, i.e. the collection of individual objects repeatedly observed at multiple time-points. The method allows to compute an average spatiotemporal trajectory of shape changes at the group level, and the individual variations of this trajectory both in terms of geometry and time dynamics. First, we formulate a non-linear mixed-effects statistical model as the combination of a generic statistical model for manifold-valued longitudinal data, a deformation model defining shape trajectories via the action of a finite-dimensional set of diffeomorphisms with a manifold structure, and an efficient numerical scheme to compute parallel transport on this manifold. Second, we introduce a MCMC-SAEM algorithm with a specific approach to shape sampling, an adaptive scheme for proposal variances, and a log-likelihood tempering strategy to estimate our model. Third, we validate our algorithm on 2D simulated data, and then estimate a scenario of alteration of the shape of the hippocampus 3D brain structure during the course of Alzheimer's disease. The method shows for instance that hippocampal atrophy progresses more quickly in female subjects, and occurs earlier in APOE4 mutation carriers. We finally illustrate the potential of our method for classifying pathological trajectories versus normal ageing.
Learning Myelin Content in Multiple Sclerosis from Multimodal MRI through Adversarial Training
Jun 08, 2018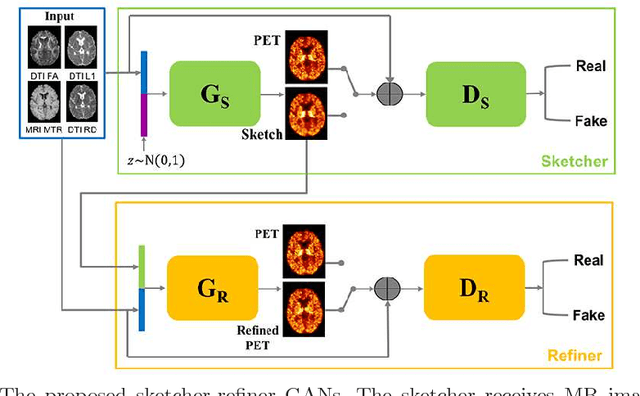
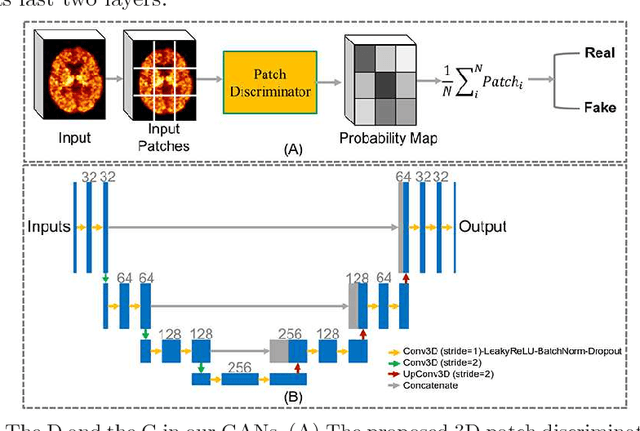
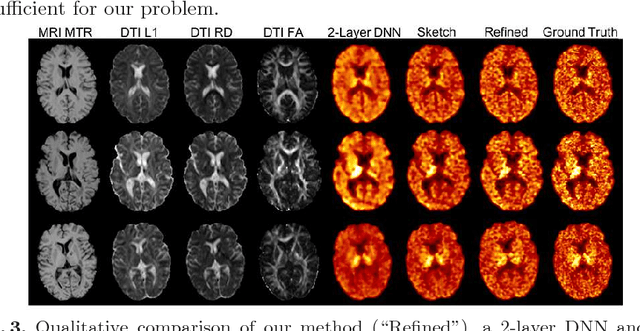
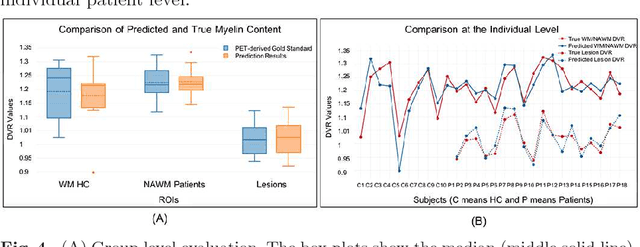
Abstract:Multiple sclerosis (MS) is a demyelinating disease of the central nervous system (CNS). A reliable measure of the tissue myelin content is therefore essential for the understanding of the physiopathology of MS, tracking progression and assessing treatment efficacy. Positron emission tomography (PET) with $[^{11} \mbox{C}] \mbox{PIB}$ has been proposed as a promising biomarker for measuring myelin content changes in-vivo in MS. However, PET imaging is expensive and invasive due to the injection of a radioactive tracer. On the contrary, magnetic resonance imaging (MRI) is a non-invasive, widely available technique, but existing MRI sequences do not provide, to date, a reliable, specific, or direct marker of either demyelination or remyelination. In this work, we therefore propose Sketcher-Refiner Generative Adversarial Networks (GANs) with specifically designed adversarial loss functions to predict the PET-derived myelin content map from a combination of MRI modalities. The prediction problem is solved by a sketch-refinement process in which the sketcher generates the preliminary anatomical and physiological information and the refiner refines and generates images reflecting the tissue myelin content in the human brain. We evaluated the ability of our method to predict myelin content at both global and voxel-wise levels. The evaluation results show that the demyelination in lesion regions and myelin content in normal-appearing white matter (NAWM) can be well predicted by our method. The method has the potential to become a useful tool for clinical management of patients with MS.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge