Igor Koval
ARAMIS, CMAP, ICM
Simulation of virtual cohorts increases predictive accuracy of cognitive decline in MCI subjects
Apr 05, 2019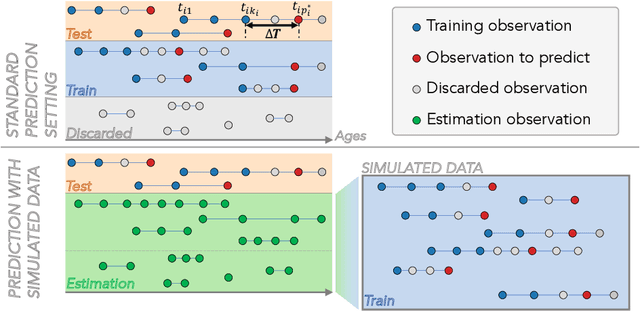
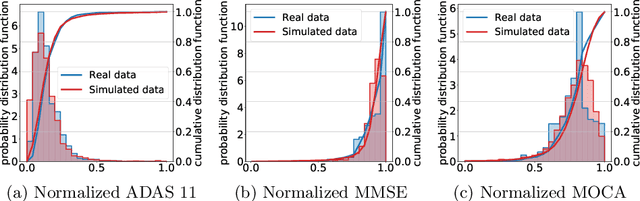
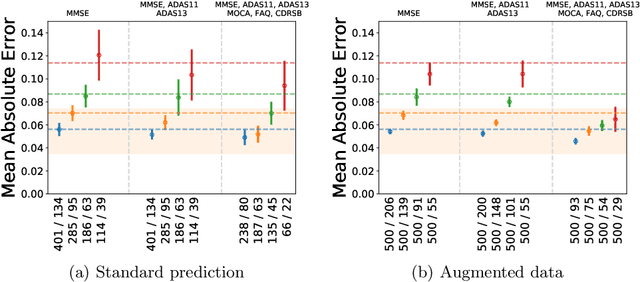
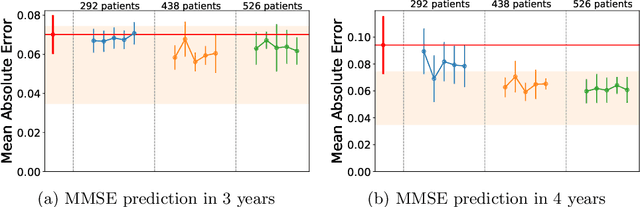
Abstract:The ability to predict the progression of biomarkers, notably in NDD, is limited by the size of the longitudinal data sets, in terms of number of patients, number of visits per patients and total follow-up time. To this end, we introduce a data augmentation technique that is able to reproduce the variability seen in a longitudinal training data set and simulate continuous biomarkers trajectories for any number of virtual patients. Thanks to this simulation framework, we propose to transform the training set into a simulated data set with more patients, more time-points per patient and longer follow-up duration. We illustrate this approach on the prediction of the MMSE of MCI subjects of the ADNI data set. We show that it allows to reach predictions with errors comparable to the noise in the data, estimated in test/retest studies, achieving a improvement of 37% of the mean absolute error compared to the same non-augmented model.
Statistical learning of spatiotemporal patterns from longitudinal manifold-valued networks
Sep 25, 2017


Abstract:We introduce a mixed-effects model to learn spatiotempo-ral patterns on a network by considering longitudinal measures distributed on a fixed graph. The data come from repeated observations of subjects at different time points which take the form of measurement maps distributed on a graph such as an image or a mesh. The model learns a typical group-average trajectory characterizing the propagation of measurement changes across the graph nodes. The subject-specific trajectories are defined via spatial and temporal transformations of the group-average scenario, thus estimating the variability of spatiotemporal patterns within the group. To estimate population and individual model parameters, we adapted a stochastic version of the Expectation-Maximization algorithm, the MCMC-SAEM. The model is used to describe the propagation of cortical atrophy during the course of Alzheimer's Disease. Model parameters show the variability of this average pattern of atrophy in terms of trajectories across brain regions, age at disease onset and pace of propagation. We show that the personalization of this model yields accurate prediction of maps of cortical thickness in patients.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge