Alexandre Routier
for the Alzheimers Disease Neuroimaging Initiative
Convolutional Neural Networks for Classification of Alzheimer's Disease: Overview and Reproducible Evaluation
Apr 16, 2019
Abstract:In the past two years, over 30 papers have proposed to use convolutional neural network (CNN) for AD classification. However, the classification performances across studies are difficult to compare. Moreover, these studies are hardly reproducible because their frameworks are not publicly accessible. Lastly, some of these papers may reported biased performances due to inadequate or unclear validation procedure and also it is unclear how the model architecture and parameters were chosen. In the present work, we aim to address these limitations through three main contributions. First, we performed a systematic literature review of studies using CNN for AD classification from anatomical MRI. We identified four main types of approaches: 2D slice-level, 3D patch-level, ROI-based and 3D subject-level CNN. Moreover, we found that more than half of the surveyed papers may have suffered from data leakage and thus reported biased performances. Our second contribution is an open-source framework for classification of AD. Thirdly, we used this framework to rigorously compare different CNN architectures, which are representative of the existing literature, and to study the influence of key components on classification performances. On the validation set, the ROI-based (hippocampus) CNN achieved highest balanced accuracy (0.86 for AD vs CN and 0.80 for sMCI vs pMCI) compared to other approaches. Transfer learning with autoencoder pre-training did not improve the average accuracy but reduced the variance. Training using longitudinal data resulted in similar or higher performance, depending on the approach, compared to training with only baseline data. Sophisticated image preprocessing did not improve the results. Lastly, CNN performed similarly to standard SVM for task AD vs CN but outperformed SVM for task sMCI vs pMCI, demonstrating the potential of deep learning for challenging diagnostic tasks.
Reproducible evaluation of diffusion MRI features for automatic classification of patients with Alzheimers disease
Dec 28, 2018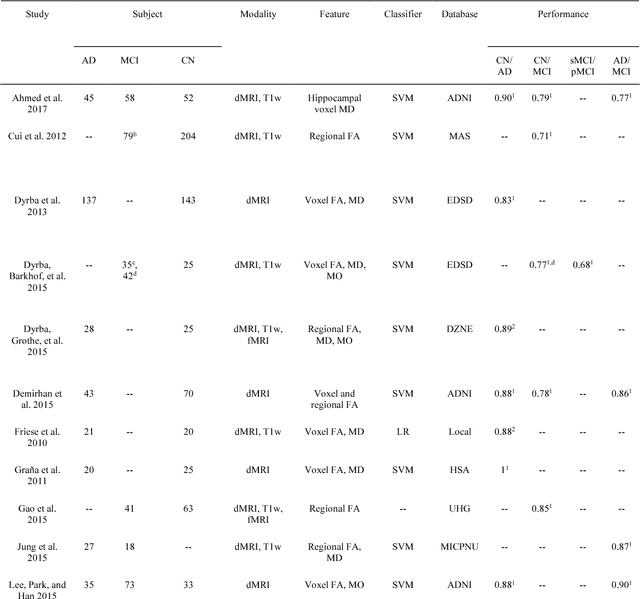
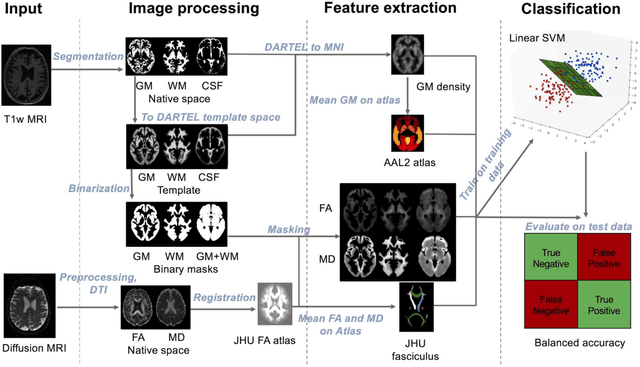
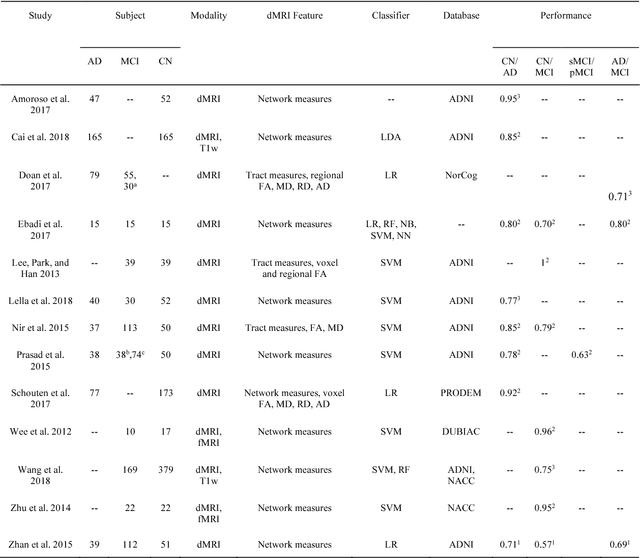
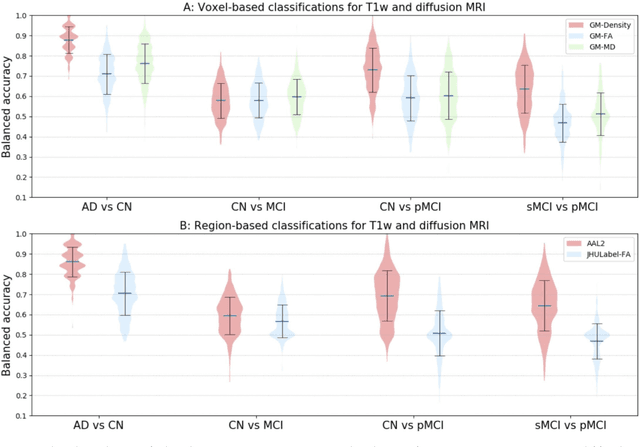
Abstract:Diffusion MRI is the modality of choice to study alterations of white matter. In the past years, various works have used diffusion MRI for automatic classification of Alzheimers disease. However, the performances obtained with different approaches are difficult to compare because of variations in components such as input data, participant selection, image preprocessing, feature extraction, feature selection (FS) and cross-validation (CV) procedure. Moreover, these studies are also difficult to reproduce because these different components are not readily available. In a previous work (Samper-Gonzalez et al. 2018), we proposed an open-source framework for the reproducible evaluation of AD classification from T1-weighted (T1w) MRI and PET data. In the present paper, we extend this framework to diffusion MRI data. The framework comprises: tools to automatically convert ADNI data into the BIDS standard, pipelines for image preprocessing and feature extraction, baseline classifiers and a rigorous CV procedure. We demonstrate the use of the framework through assessing the influence of diffusion tensor imaging (DTI) metrics (fractional anisotropy - FA, mean diffusivity - MD), feature types, imaging modalities (diffusion MRI or T1w MRI), data imbalance and FS bias. First, voxel-wise features generally gave better performances than regional features. Secondly, FA and MD provided comparable results for voxel-wise features. Thirdly, T1w MRI performed better than diffusion MRI. Fourthly, we demonstrated that using non-nested validation of FS leads to unreliable and over-optimistic results. All the code is publicly available: general-purpose tools have been integrated into the Clinica software (www.clinica.run) and the paper-specific code is available at: https://gitlab.icm-institute.org/aramislab/AD-ML.
Reproducible evaluation of classification methods in Alzheimer's disease: framework and application to MRI and PET data
Aug 20, 2018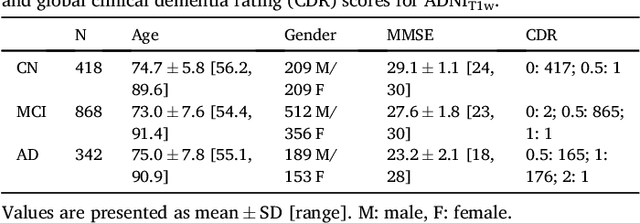
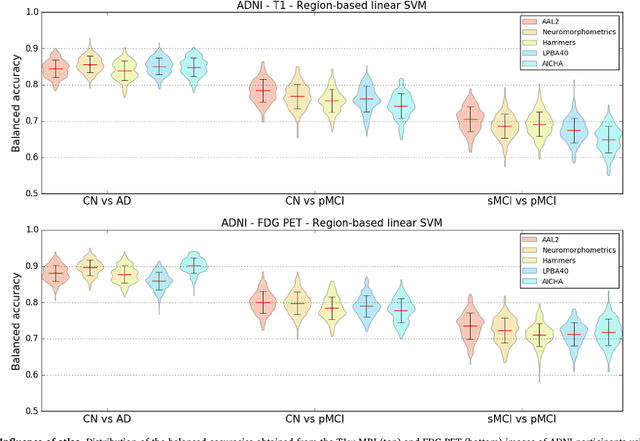
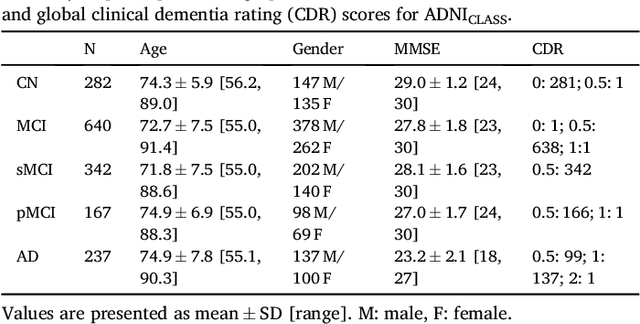
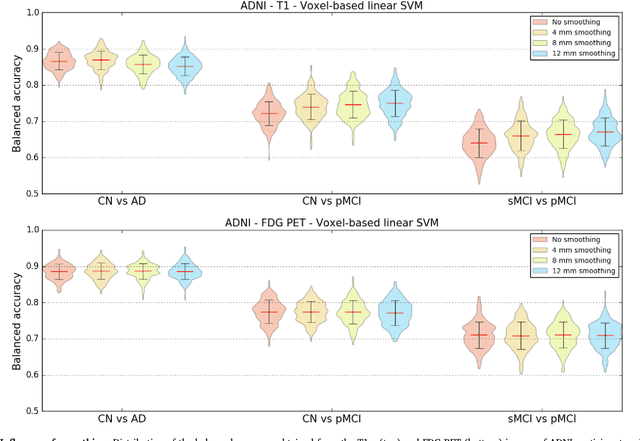
Abstract:A large number of papers have introduced novel machine learning and feature extraction methods for automatic classification of AD. However, they are difficult to reproduce because key components of the validation are often not readily available. These components include selected participants and input data, image preprocessing and cross-validation procedures. The performance of the different approaches is also difficult to compare objectively. In particular, it is often difficult to assess which part of the method provides a real improvement, if any. We propose a framework for reproducible and objective classification experiments in AD using three publicly available datasets (ADNI, AIBL and OASIS). The framework comprises: i) automatic conversion of the three datasets into BIDS format, ii) a modular set of preprocessing pipelines, feature extraction and classification methods, together with an evaluation framework, that provide a baseline for benchmarking the different components. We demonstrate the use of the framework for a large-scale evaluation on 1960 participants using T1 MRI and FDG PET data. In this evaluation, we assess the influence of different modalities, preprocessing, feature types, classifiers, training set sizes and datasets. Performances were in line with the state-of-the-art. FDG PET outperformed T1 MRI for all classification tasks. No difference in performance was found for the use of different atlases, image smoothing, partial volume correction of FDG PET images, or feature type. Linear SVM and L2-logistic regression resulted in similar performance and both outperformed random forests. The classification performance increased along with the number of subjects used for training. Classifiers trained on ADNI generalized well to AIBL and OASIS. All the code of the framework and the experiments is publicly available at: https://gitlab.icm-institute.org/aramislab/AD-ML.
Prediction of the progression of subcortical brain structures in Alzheimer's disease from baseline
Nov 23, 2017


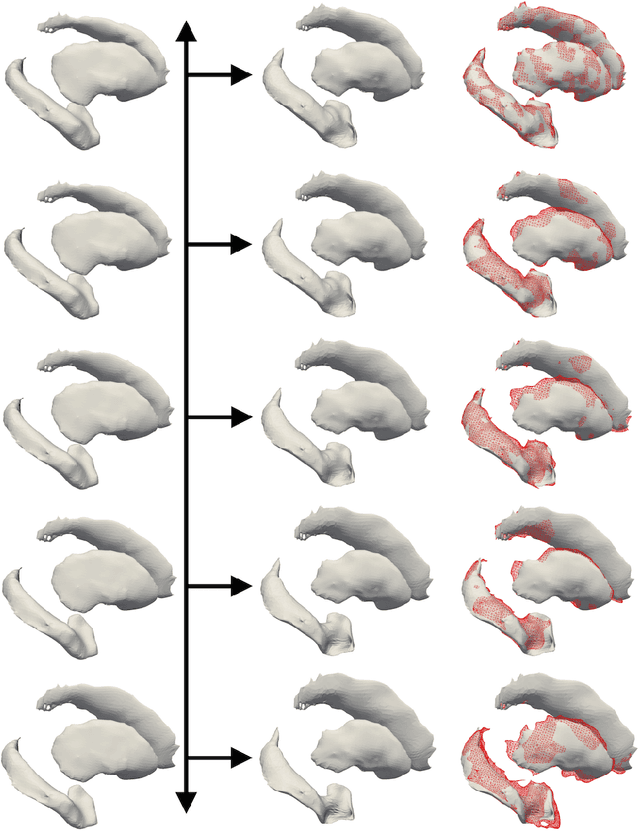
Abstract:We propose a method to predict the subject-specific longitudinal progression of brain structures extracted from baseline MRI, and evaluate its performance on Alzheimer's disease data. The disease progression is modeled as a trajectory on a group of diffeomorphisms in the context of large deformation diffeomorphic metric mapping (LDDMM). We first exhibit the limited predictive abilities of geodesic regression extrapolation on this group. Building on the recent concept of parallel curves in shape manifolds, we then introduce a second predictive protocol which personalizes previously learned trajectories to new subjects, and investigate the relative performances of two parallel shifting paradigms. This design only requires the baseline imaging data. Finally, coefficients encoding the disease dynamics are obtained from longitudinal cognitive measurements for each subject, and exploited to refine our methodology which is demonstrated to successfully predict the follow-up visits.
Statistical learning of spatiotemporal patterns from longitudinal manifold-valued networks
Sep 25, 2017


Abstract:We introduce a mixed-effects model to learn spatiotempo-ral patterns on a network by considering longitudinal measures distributed on a fixed graph. The data come from repeated observations of subjects at different time points which take the form of measurement maps distributed on a graph such as an image or a mesh. The model learns a typical group-average trajectory characterizing the propagation of measurement changes across the graph nodes. The subject-specific trajectories are defined via spatial and temporal transformations of the group-average scenario, thus estimating the variability of spatiotemporal patterns within the group. To estimate population and individual model parameters, we adapted a stochastic version of the Expectation-Maximization algorithm, the MCMC-SAEM. The model is used to describe the propagation of cortical atrophy during the course of Alzheimer's Disease. Model parameters show the variability of this average pattern of atrophy in terms of trajectories across brain regions, age at disease onset and pace of propagation. We show that the personalization of this model yields accurate prediction of maps of cortical thickness in patients.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge