Stephane Epelbaum
for the Alzheimers Disease Neuroimaging Initiative
Reproducible evaluation of diffusion MRI features for automatic classification of patients with Alzheimers disease
Dec 28, 2018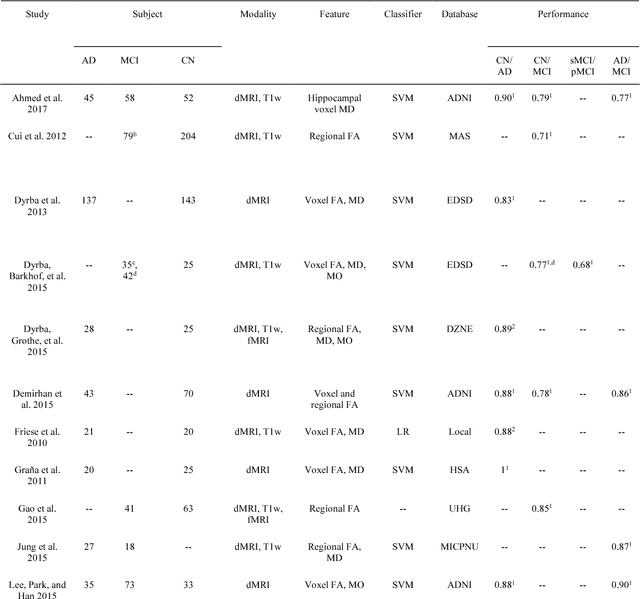
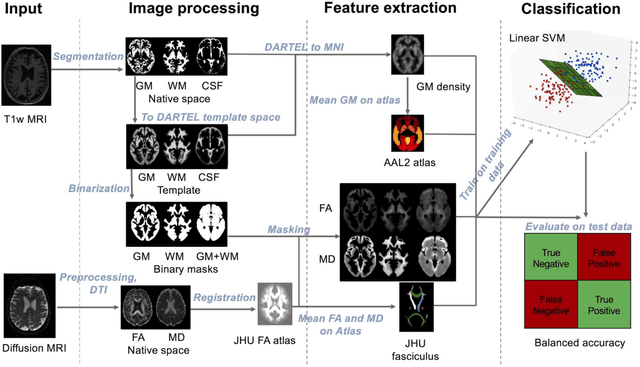
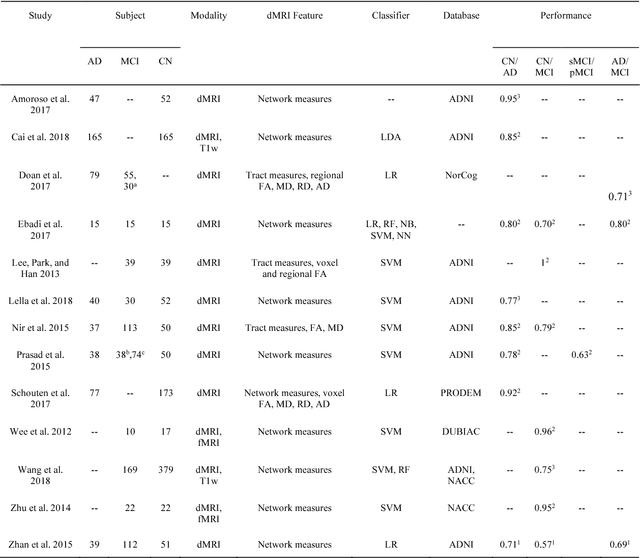
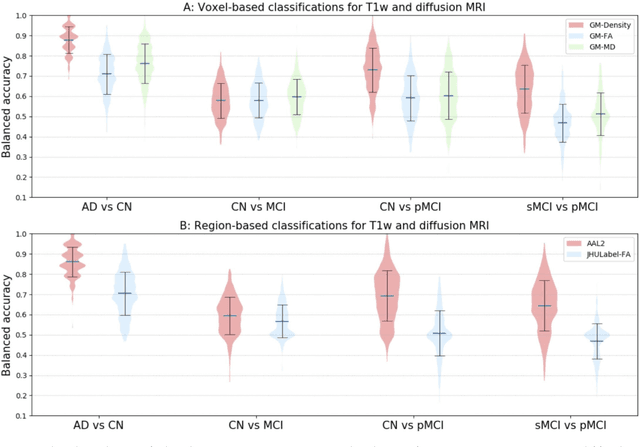
Abstract:Diffusion MRI is the modality of choice to study alterations of white matter. In the past years, various works have used diffusion MRI for automatic classification of Alzheimers disease. However, the performances obtained with different approaches are difficult to compare because of variations in components such as input data, participant selection, image preprocessing, feature extraction, feature selection (FS) and cross-validation (CV) procedure. Moreover, these studies are also difficult to reproduce because these different components are not readily available. In a previous work (Samper-Gonzalez et al. 2018), we proposed an open-source framework for the reproducible evaluation of AD classification from T1-weighted (T1w) MRI and PET data. In the present paper, we extend this framework to diffusion MRI data. The framework comprises: tools to automatically convert ADNI data into the BIDS standard, pipelines for image preprocessing and feature extraction, baseline classifiers and a rigorous CV procedure. We demonstrate the use of the framework through assessing the influence of diffusion tensor imaging (DTI) metrics (fractional anisotropy - FA, mean diffusivity - MD), feature types, imaging modalities (diffusion MRI or T1w MRI), data imbalance and FS bias. First, voxel-wise features generally gave better performances than regional features. Secondly, FA and MD provided comparable results for voxel-wise features. Thirdly, T1w MRI performed better than diffusion MRI. Fourthly, we demonstrated that using non-nested validation of FS leads to unreliable and over-optimistic results. All the code is publicly available: general-purpose tools have been integrated into the Clinica software (www.clinica.run) and the paper-specific code is available at: https://gitlab.icm-institute.org/aramislab/AD-ML.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge