Maxime Louis
DiffLoRA: Differential Low-Rank Adapters for Large Language Models
Jul 31, 2025Abstract:Differential Transformer has recently been proposed to improve performance in Transformer models by canceling out noise through a denoiser attention mechanism. In this work, we introduce DiffLoRA, a parameter-efficient adaptation of the differential attention mechanism, with low-rank adapters on both positive and negative attention terms. This approach retains the efficiency of LoRA while aiming to benefit from the performance gains of differential attention. We evaluate DiffLoRA across a broad range of NLP tasks, including general benchmarks, many-shot in-context learning, RAG, and long-context tests. We observe that, although DiffLoRA falls short of other parameter-efficient fine-tuning methods in most evaluation tasks, it shows interesting results in certain domains (+11 pts on LoRA for HumanEval). We analyze the attention patterns post-finetuning to identify the reasons for this behavior.
Adapting Large Language Models for Multi-Domain Retrieval-Augmented-Generation
Apr 03, 2025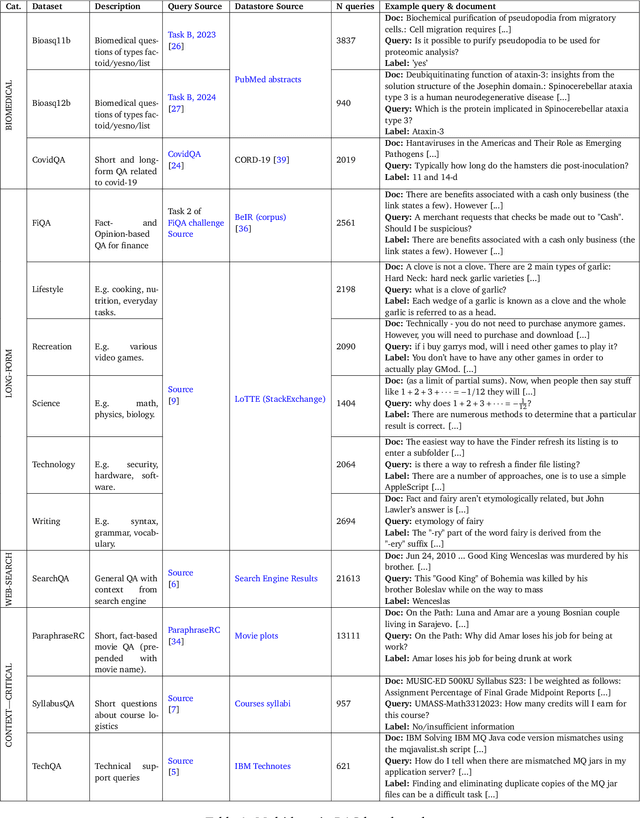
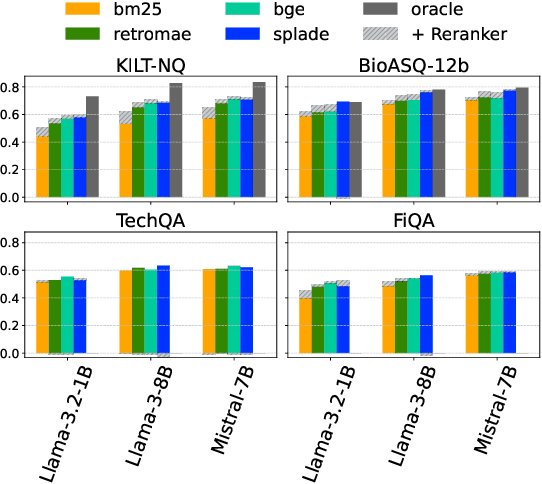
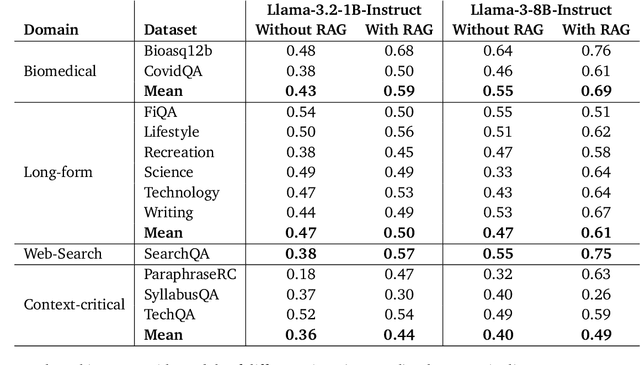
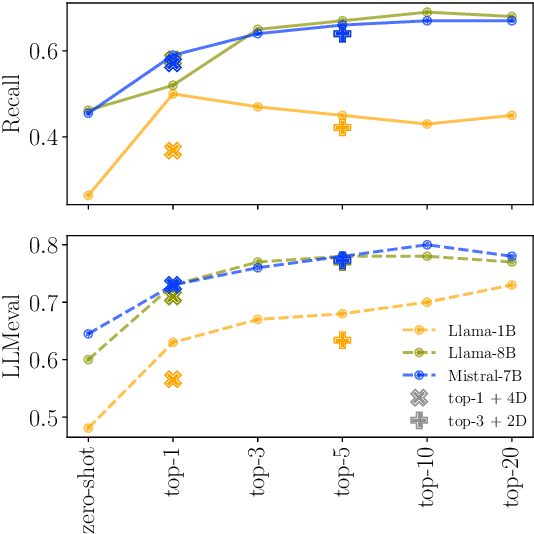
Abstract:Retrieval-Augmented Generation (RAG) enhances LLM factuality, but multi-domain applications face challenges like lack of diverse benchmarks and poor out-of-domain generalization. The first contribution of this work is to introduce a diverse benchmark comprising a variety of question-answering tasks from 8 sources and covering 13 domains. Our second contribution consists in systematically testing out-of-domain generalization for typical RAG tuning strategies. While our findings reveal that standard fine-tuning fails to generalize effectively, we show that sequence-level distillation with teacher-generated labels improves out-of-domain performance by providing more coherent supervision. Our findings highlight key strategies for improving multi-domain RAG robustness.
PISCO: Pretty Simple Compression for Retrieval-Augmented Generation
Jan 27, 2025Abstract:Retrieval-Augmented Generation (RAG) pipelines enhance Large Language Models (LLMs) by retrieving relevant documents, but they face scalability issues due to high inference costs and limited context size. Document compression is a practical solution, but current soft compression methods suffer from accuracy losses and require extensive pretraining. In this paper, we introduce PISCO, a novel method that achieves a 16x compression rate with minimal accuracy loss (0-3%) across diverse RAG-based question-answering (QA) tasks. Unlike existing approaches, PISCO requires no pretraining or annotated data, relying solely on sequence-level knowledge distillation from document-based questions. With the ability to fine-tune a 7-10B LLM in 48 hours on a single A100 GPU, PISCO offers a highly efficient and scalable solution. We present comprehensive experiments showing that PISCO outperforms existing compression models by 8% in accuracy.
End-to-end Offline Reinforcement Learning for Glycemia Control
Oct 16, 2023



Abstract:The development of closed-loop systems for glycemia control in type I diabetes relies heavily on simulated patients. Improving the performances and adaptability of these close-loops raises the risk of over-fitting the simulator. This may have dire consequences, especially in unusual cases which were not faithfully-if at all-captured by the simulator. To address this, we propose to use offline RL agents, trained on real patient data, to perform the glycemia control. To further improve the performances, we propose an end-to-end personalization pipeline, which leverages offline-policy evaluation methods to remove altogether the need of a simulator, while still enabling an estimation of clinically relevant metrics for diabetes.
Leveraging 3D Information in Unsupervised Brain MRI Segmentation
Jan 26, 2021

Abstract:Automatic segmentation of brain abnormalities is challenging, as they vary considerably from one pathology to another. Current methods are supervised and require numerous annotated images for each pathology, a strenuous task. To tackle anatomical variability, Unsupervised Anomaly Detection (UAD) methods are proposed, detecting anomalies as outliers of a healthy model learned using a Variational Autoencoder (VAE). Previous work on UAD adopted a 2D approach, meaning that MRIs are processed as a collection of independent slices. Yet, it does not fully exploit the spatial information contained in MRI. Here, we propose to perform UAD in a 3D fashion and compare 2D and 3D VAEs. As a side contribution, we present a new loss function guarantying a robust training. Learning is performed using a multicentric dataset of healthy brain MRIs, and segmentation performances are estimated on White-Matter Hyperintensities and tumors lesions. Experiments demonstrate the interest of 3D methods which outperform their 2D counterparts.
Parallel transport in shape analysis: a scalable numerical scheme
Nov 23, 2017



Abstract:The analysis of manifold-valued data requires efficient tools from Riemannian geometry to cope with the computational complexity at stake. This complexity arises from the always-increasing dimension of the data, and the absence of closed-form expressions to basic operations such as the Riemannian logarithm. In this paper, we adapt a generic numerical scheme recently introduced for computing parallel transport along geodesics in a Riemannian manifold to finite-dimensional manifolds of diffeomorphisms. We provide a qualitative and quantitative analysis of its behavior on high-dimensional manifolds, and investigate an application with the prediction of brain structures progression.
Prediction of the progression of subcortical brain structures in Alzheimer's disease from baseline
Nov 23, 2017


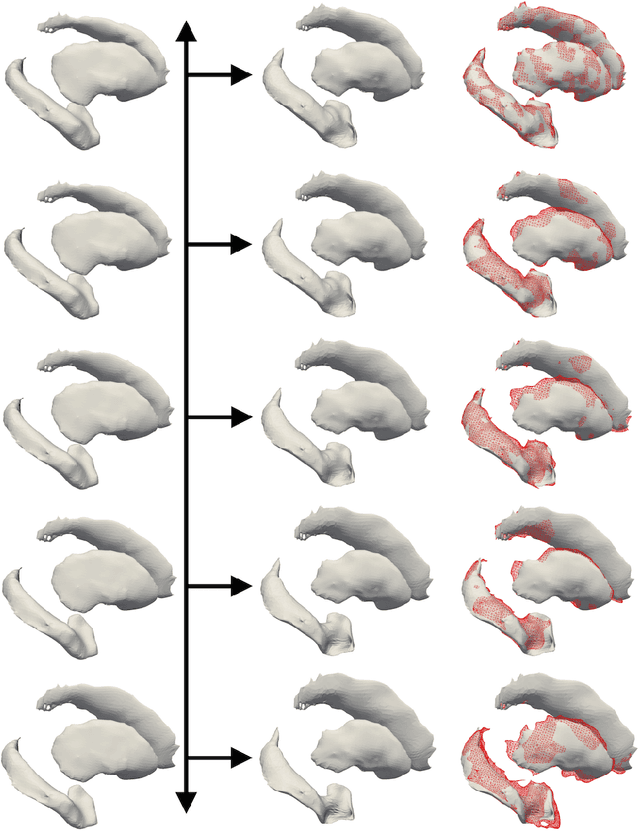
Abstract:We propose a method to predict the subject-specific longitudinal progression of brain structures extracted from baseline MRI, and evaluate its performance on Alzheimer's disease data. The disease progression is modeled as a trajectory on a group of diffeomorphisms in the context of large deformation diffeomorphic metric mapping (LDDMM). We first exhibit the limited predictive abilities of geodesic regression extrapolation on this group. Building on the recent concept of parallel curves in shape manifolds, we then introduce a second predictive protocol which personalizes previously learned trajectories to new subjects, and investigate the relative performances of two parallel shifting paradigms. This design only requires the baseline imaging data. Finally, coefficients encoding the disease dynamics are obtained from longitudinal cognitive measurements for each subject, and exploited to refine our methodology which is demonstrated to successfully predict the follow-up visits.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge