Benjamin Lambert
Robust Conformal Volume Estimation in 3D Medical Images
Jul 29, 2024



Abstract:Volumetry is one of the principal downstream applications of 3D medical image segmentation, for example, to detect abnormal tissue growth or for surgery planning. Conformal Prediction is a promising framework for uncertainty quantification, providing calibrated predictive intervals associated with automatic volume measurements. However, this methodology is based on the hypothesis that calibration and test samples are exchangeable, an assumption that is in practice often violated in medical image applications. A weighted formulation of Conformal Prediction can be framed to mitigate this issue, but its empirical investigation in the medical domain is still lacking. A potential reason is that it relies on the estimation of the density ratio between the calibration and test distributions, which is likely to be intractable in scenarios involving high-dimensional data. To circumvent this, we propose an efficient approach for density ratio estimation relying on the compressed latent representations generated by the segmentation model. Our experiments demonstrate the efficiency of our approach to reduce the coverage error in the presence of covariate shifts, in both synthetic and real-world settings. Our implementation is available at https://github.com/benolmbrt/wcp_miccai
Anisotropic Hybrid Networks for liver tumor segmentation with uncertainty quantification
Aug 23, 2023



Abstract:The burden of liver tumors is important, ranking as the fourth leading cause of cancer mortality. In case of hepatocellular carcinoma (HCC), the delineation of liver and tumor on contrast-enhanced magnetic resonance imaging (CE-MRI) is performed to guide the treatment strategy. As this task is time-consuming, needs high expertise and could be subject to inter-observer variability there is a strong need for automatic tools. However, challenges arise from the lack of available training data, as well as the high variability in terms of image resolution and MRI sequence. In this work we propose to compare two different pipelines based on anisotropic models to obtain the segmentation of the liver and tumors. The first pipeline corresponds to a baseline multi-class model that performs the simultaneous segmentation of the liver and tumor classes. In the second approach, we train two distinct binary models, one segmenting the liver only and the other the tumors. Our results show that both pipelines exhibit different strengths and weaknesses. Moreover we propose an uncertainty quantification strategy allowing the identification of potential false positive tumor lesions. Both solutions were submitted to the MICCAI 2023 Atlas challenge regarding liver and tumor segmentation.
Multi-layer Aggregation as a key to feature-based OOD detection
Jul 28, 2023Abstract:Deep Learning models are easily disturbed by variations in the input images that were not observed during the training stage, resulting in unpredictable predictions. Detecting such Out-of-Distribution (OOD) images is particularly crucial in the context of medical image analysis, where the range of possible abnormalities is extremely wide. Recently, a new category of methods has emerged, based on the analysis of the intermediate features of a trained model. These methods can be divided into 2 groups: single-layer methods that consider the feature map obtained at a fixed, carefully chosen layer, and multi-layer methods that consider the ensemble of the feature maps generated by the model. While promising, a proper comparison of these algorithms is still lacking. In this work, we compared various feature-based OOD detection methods on a large spectra of OOD (20 types), representing approximately 7800 3D MRIs. Our experiments shed the light on two phenomenons. First, multi-layer methods consistently outperform single-layer approaches, which tend to have inconsistent behaviour depending on the type of anomaly. Second, the OOD detection performance highly depends on the architecture of the underlying neural network.
TriadNet: Sampling-free predictive intervals for lesional volume in 3D brain MR images
Jul 28, 2023Abstract:The volume of a brain lesion (e.g. infarct or tumor) is a powerful indicator of patient prognosis and can be used to guide the therapeutic strategy. Lesional volume estimation is usually performed by segmentation with deep convolutional neural networks (CNN), currently the state-of-the-art approach. However, to date, few work has been done to equip volume segmentation tools with adequate quantitative predictive intervals, which can hinder their usefulness and acceptation in clinical practice. In this work, we propose TriadNet, a segmentation approach relying on a multi-head CNN architecture, which provides both the lesion volumes and the associated predictive intervals simultaneously, in less than a second. We demonstrate its superiority over other solutions on BraTS 2021, a large-scale MRI glioblastoma image database.
Improving Uncertainty-based Out-of-Distribution Detection for Medical Image Segmentation
Nov 10, 2022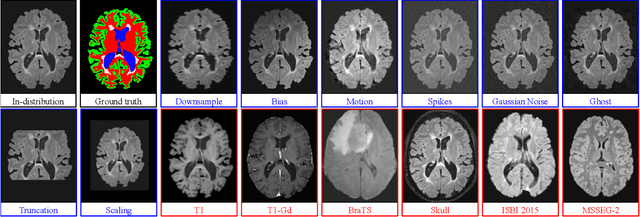
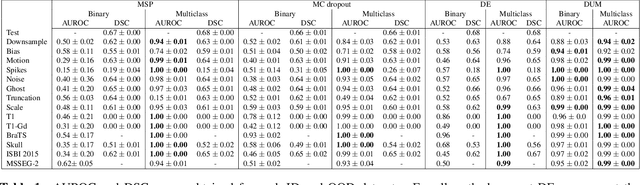
Abstract:Deep Learning models are easily disturbed by variations in the input images that were not seen during training, resulting in unpredictable behaviours. Such Out-of-Distribution (OOD) images represent a significant challenge in the context of medical image analysis, where the range of possible abnormalities is extremely wide, including artifacts, unseen pathologies, or different imaging protocols. In this work, we evaluate various uncertainty frameworks to detect OOD inputs in the context of Multiple Sclerosis lesions segmentation. By implementing a comprehensive evaluation scheme including 14 sources of OOD of various nature and strength, we show that methods relying on the predictive uncertainty of binary segmentation models often fails in detecting outlying inputs. On the contrary, learning to segment anatomical labels alongside lesions highly improves the ability to detect OOD inputs.
Trustworthy clinical AI solutions: a unified review of uncertainty quantification in deep learning models for medical image analysis
Oct 05, 2022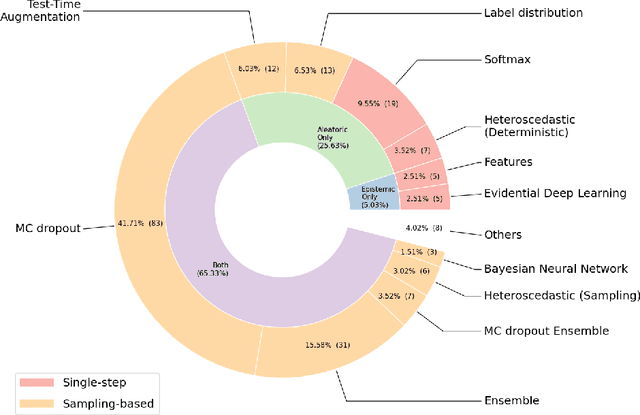
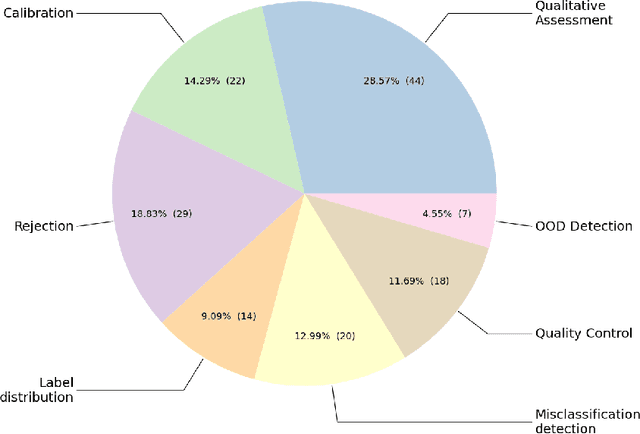
Abstract:The full acceptance of Deep Learning (DL) models in the clinical field is rather low with respect to the quantity of high-performing solutions reported in the literature. Particularly, end users are reluctant to rely on the rough predictions of DL models. Uncertainty quantification methods have been proposed in the literature as a potential response to reduce the rough decision provided by the DL black box and thus increase the interpretability and the acceptability of the result by the final user. In this review, we propose an overview of the existing methods to quantify uncertainty associated to DL predictions. We focus on applications to medical image analysis, which present specific challenges due to the high dimensionality of images and their quality variability, as well as constraints associated to real-life clinical routine. We then discuss the evaluation protocols to validate the relevance of uncertainty estimates. Finally, we highlight the open challenges of uncertainty quantification in the medical field.
Beyond Voxel Prediction Uncertainty: Identifying brain lesions you can trust
Sep 22, 2022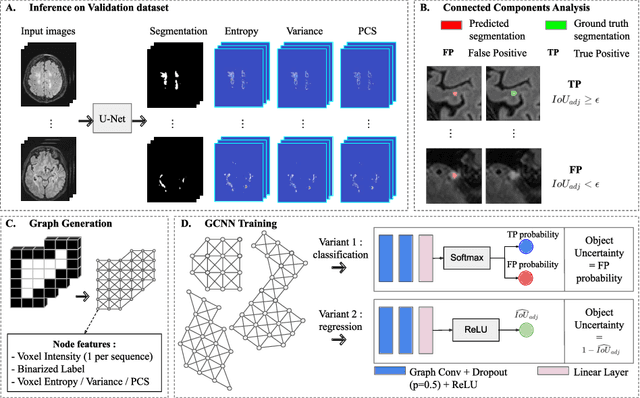
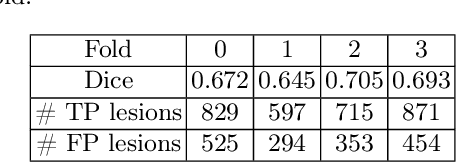
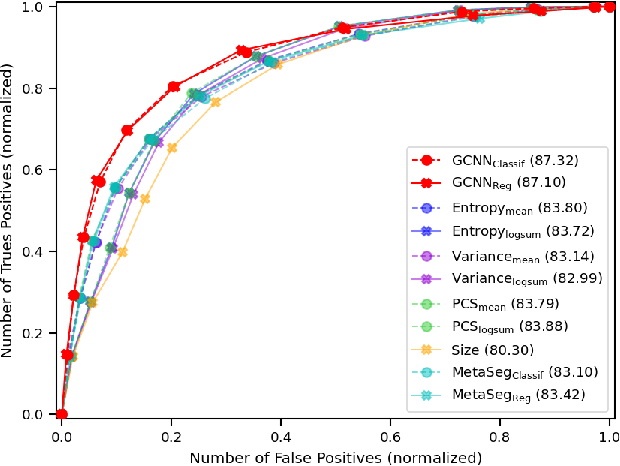
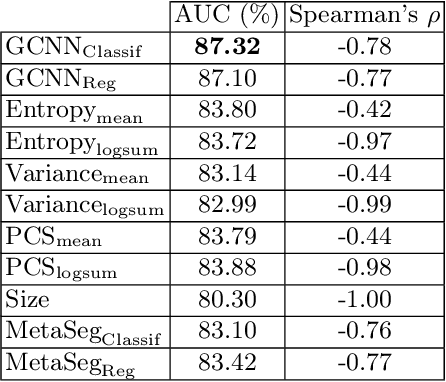
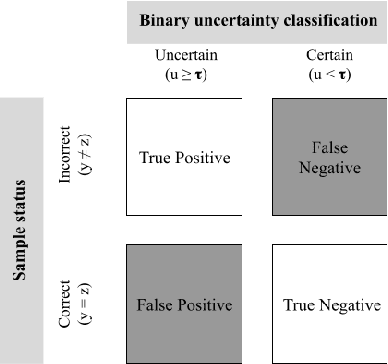
Abstract:Deep neural networks have become the gold-standard approach for the automated segmentation of 3D medical images. Their full acceptance by clinicians remains however hampered by the lack of intelligible uncertainty assessment of the provided results. Most approaches to quantify their uncertainty, such as the popular Monte Carlo dropout, restrict to some measure of uncertainty in prediction at the voxel level. In addition not to be clearly related to genuine medical uncertainty, this is not clinically satisfying as most objects of interest (e.g. brain lesions) are made of groups of voxels whose overall relevance may not simply reduce to the sum or mean of their individual uncertainties. In this work, we propose to go beyond voxel-wise assessment using an innovative Graph Neural Network approach, trained from the outputs of a Monte Carlo dropout model. This network allows the fusion of three estimators of voxel uncertainty: entropy, variance, and model's confidence; and can be applied to any lesion, regardless of its shape or size. We demonstrate the superiority of our approach for uncertainty estimate on a task of Multiple Sclerosis lesions segmentation.
Leveraging 3D Information in Unsupervised Brain MRI Segmentation
Jan 26, 2021

Abstract:Automatic segmentation of brain abnormalities is challenging, as they vary considerably from one pathology to another. Current methods are supervised and require numerous annotated images for each pathology, a strenuous task. To tackle anatomical variability, Unsupervised Anomaly Detection (UAD) methods are proposed, detecting anomalies as outliers of a healthy model learned using a Variational Autoencoder (VAE). Previous work on UAD adopted a 2D approach, meaning that MRIs are processed as a collection of independent slices. Yet, it does not fully exploit the spatial information contained in MRI. Here, we propose to perform UAD in a 3D fashion and compare 2D and 3D VAEs. As a side contribution, we present a new loss function guarantying a robust training. Learning is performed using a multicentric dataset of healthy brain MRIs, and segmentation performances are estimated on White-Matter Hyperintensities and tumors lesions. Experiments demonstrate the interest of 3D methods which outperform their 2D counterparts.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge