Elina Thibeau-Sutre
Brain-Shift: Unsupervised Pseudo-Healthy Brain Synthesis for Novel Biomarker Extraction in Chronic Subdural Hematoma
Mar 28, 2024



Abstract:Chronic subdural hematoma (cSDH) is a common neurological condition characterized by the accumulation of blood between the brain and the dura mater. This accumulation of blood can exert pressure on the brain, potentially leading to fatal outcomes. Treatment options for cSDH are limited to invasive surgery or non-invasive management. Traditionally, the midline shift, hand-measured by experts from an ideal sagittal plane, and the hematoma volume have been the primary metrics for quantifying and analyzing cSDH. However, these approaches do not quantify the local 3D brain deformation caused by cSDH. We propose a novel method using anatomy-aware unsupervised diffeomorphic pseudo-healthy synthesis to generate brain deformation fields. The deformation fields derived from this process are utilized to extract biomarkers that quantify the shift in the brain due to cSDH. We use CT scans of 121 patients for training and validation of our method and find that our metrics allow the identification of patients who require surgery. Our results indicate that automatically obtained brain deformation fields might contain prognostic value for personalized cSDH treatment. Our implementation is available on: github.com/Barisimre/brain-morphing
Uncertainty-based quality assurance of carotid artery wall segmentation in black-blood MRI
Aug 18, 2023Abstract:The application of deep learning models to large-scale data sets requires means for automatic quality assurance. We have previously developed a fully automatic algorithm for carotid artery wall segmentation in black-blood MRI that we aim to apply to large-scale data sets. This method identifies nested artery walls in 3D patches centered on the carotid artery. In this study, we investigate to what extent the uncertainty in the model predictions for the contour location can serve as a surrogate for error detection and, consequently, automatic quality assurance. We express the quality of automatic segmentations using the Dice similarity coefficient. The uncertainty in the model's prediction is estimated using either Monte Carlo dropout or test-time data augmentation. We found that (1) including uncertainty measurements did not degrade the quality of the segmentations, (2) uncertainty metrics provide a good proxy of the quality of our contours if the center found during the first step is enclosed in the lumen of the carotid artery and (3) they could be used to detect low-quality segmentations at the participant level. This automatic quality assurance tool might enable the application of our model in large-scale data sets.
Reproducibility in machine learning for medical imaging
Sep 12, 2022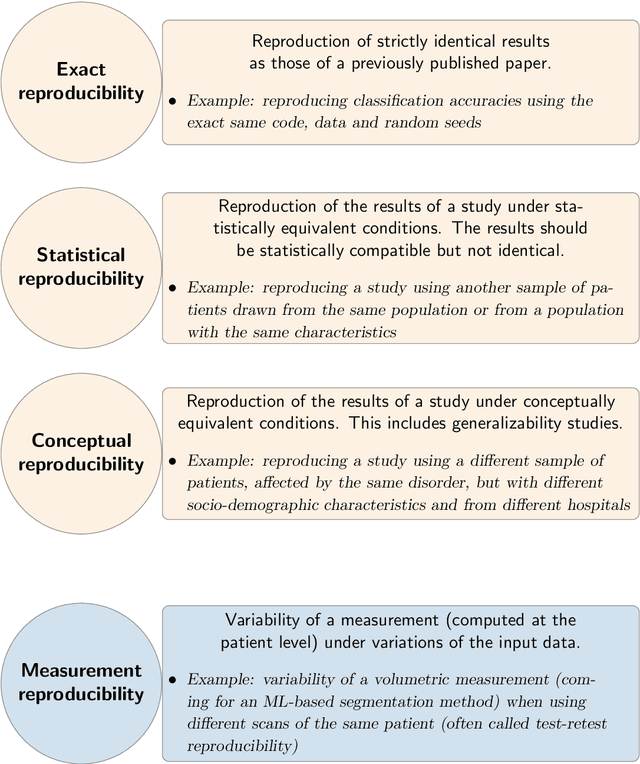
Abstract:Reproducibility is a cornerstone of science, as the replication of findings is the process through which they become knowledge. It is widely considered that many fields of science are undergoing a reproducibility crisis. This has led to the publications of various guidelines in order to improve research reproducibility. This didactic chapter intends at being an introduction to reproducibility for researchers in the field of machine learning for medical imaging. We first distinguish between different types of reproducibility. For each of them, we aim at defining it, at describing the requirements to achieve it and at discussing its utility. The chapter ends with a discussion on the benefits of reproducibility and with a plea for a non-dogmatic approach to this concept and its implementation in research practice.
Interpretability of Machine Learning Methods Applied to Neuroimaging
Apr 14, 2022



Abstract:Deep learning methods have become very popular for the processing of natural images, and were then successfully adapted to the neuroimaging field. As these methods are non-transparent, interpretability methods are needed to validate them and ensure their reliability. Indeed, it has been shown that deep learning models may obtain high performance even when using irrelevant features, by exploiting biases in the training set. Such undesirable situations can potentially be detected by using interpretability methods. Recently, many methods have been proposed to interpret neural networks. However, this domain is not mature yet. Machine learning users face two major issues when aiming to interpret their models: which method to choose, and how to assess its reliability? Here, we aim at providing answers to these questions by presenting the most common interpretability methods and metrics developed to assess their reliability, as well as their applications and benchmarks in the neuroimaging context. Note that this is not an exhaustive survey: we aimed to focus on the studies which we found to be the most representative and relevant.
Data Augmentation in High Dimensional Low Sample Size Setting Using a Geometry-Based Variational Autoencoder
Apr 30, 2021
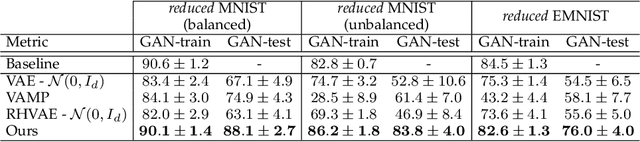

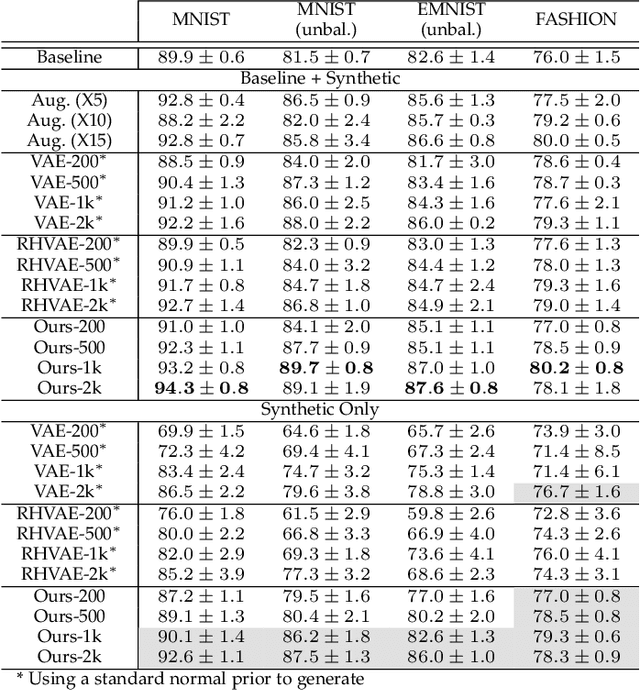
Abstract:In this paper, we propose a new method to perform data augmentation in a reliable way in the High Dimensional Low Sample Size (HDLSS) setting using a geometry-based variational autoencoder. Our approach combines a proper latent space modeling of the VAE seen as a Riemannian manifold with a new generation scheme which produces more meaningful samples especially in the context of small data sets. The proposed method is tested through a wide experimental study where its robustness to data sets, classifiers and training samples size is stressed. It is also validated on a medical imaging classification task on the challenging ADNI database where a small number of 3D brain MRIs are considered and augmented using the proposed VAE framework. In each case, the proposed method allows for a significant and reliable gain in the classification metrics. For instance, balanced accuracy jumps from 66.3% to 74.3% for a state-of-the-art CNN classifier trained with 50 MRIs of cognitively normal (CN) and 50 Alzheimer disease (AD) patients and from 77.7% to 86.3% when trained with 243 CN and 210 AD while improving greatly sensitivity and specificity metrics.
Convolutional Neural Networks for Classification of Alzheimer's Disease: Overview and Reproducible Evaluation
Apr 16, 2019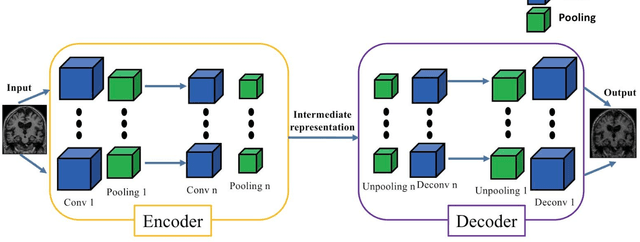
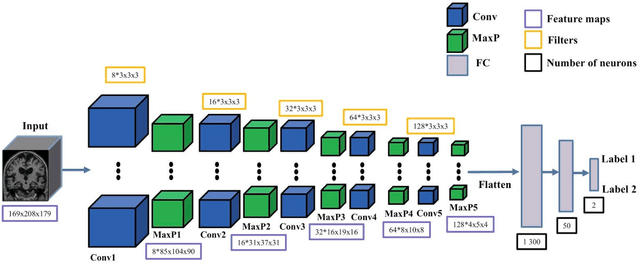
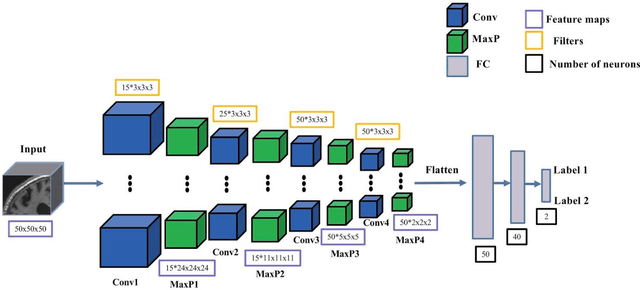
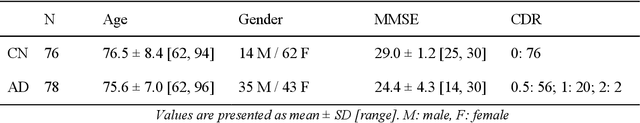
Abstract:In the past two years, over 30 papers have proposed to use convolutional neural network (CNN) for AD classification. However, the classification performances across studies are difficult to compare. Moreover, these studies are hardly reproducible because their frameworks are not publicly accessible. Lastly, some of these papers may reported biased performances due to inadequate or unclear validation procedure and also it is unclear how the model architecture and parameters were chosen. In the present work, we aim to address these limitations through three main contributions. First, we performed a systematic literature review of studies using CNN for AD classification from anatomical MRI. We identified four main types of approaches: 2D slice-level, 3D patch-level, ROI-based and 3D subject-level CNN. Moreover, we found that more than half of the surveyed papers may have suffered from data leakage and thus reported biased performances. Our second contribution is an open-source framework for classification of AD. Thirdly, we used this framework to rigorously compare different CNN architectures, which are representative of the existing literature, and to study the influence of key components on classification performances. On the validation set, the ROI-based (hippocampus) CNN achieved highest balanced accuracy (0.86 for AD vs CN and 0.80 for sMCI vs pMCI) compared to other approaches. Transfer learning with autoencoder pre-training did not improve the average accuracy but reduced the variance. Training using longitudinal data resulted in similar or higher performance, depending on the approach, compared to training with only baseline data. Sophisticated image preprocessing did not improve the results. Lastly, CNN performed similarly to standard SVM for task AD vs CN but outperformed SVM for task sMCI vs pMCI, demonstrating the potential of deep learning for challenging diagnostic tasks.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge