Alexandre Bone
Learning to diagnose cirrhosis from radiological and histological labels with joint self and weakly-supervised pretraining strategies
Feb 16, 2023Abstract:Identifying cirrhosis is key to correctly assess the health of the liver. However, the gold standard diagnosis of the cirrhosis needs a medical intervention to obtain the histological confirmation, e.g. the METAVIR score, as the radiological presentation can be equivocal. In this work, we propose to leverage transfer learning from large datasets annotated by radiologists, which we consider as a weak annotation, to predict the histological score available on a small annex dataset. To this end, we propose to compare different pretraining methods, namely weakly-supervised and self-supervised ones, to improve the prediction of the cirrhosis. Finally, we introduce a loss function combining both supervised and self-supervised frameworks for pretraining. This method outperforms the baseline classification of the METAVIR score, reaching an AUC of 0.84 and a balanced accuracy of 0.75, compared to 0.77 and 0.72 for a baseline classifier.
CoRe: An Automated Pipeline for The Prediction of Liver Resection Complexity from Preoperative CT Scans
Oct 15, 2022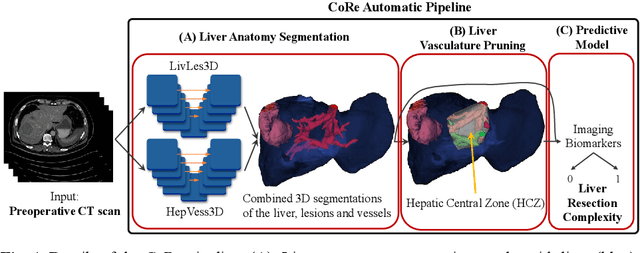
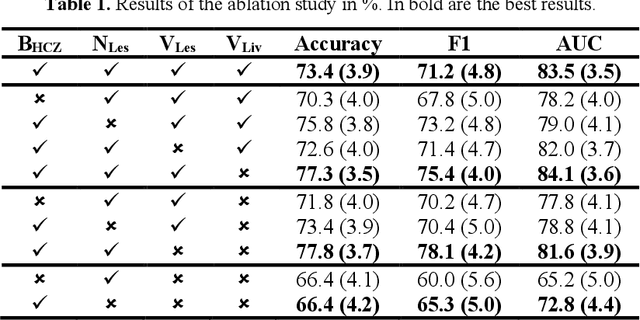
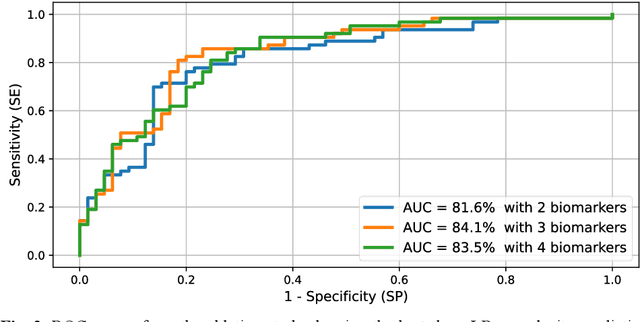

Abstract:Surgical resections are the most prevalent curative treatment for primary liver cancer. Tumors located in critical positions are known to complexify liver resections (LR). While experienced surgeons in specialized medical centers may have the necessary expertise to accurately anticipate LR complexity, and prepare accordingly, an objective method able to reproduce this behavior would have the potential to improve the standard routine of care, and avoid intra- and postoperative complications. In this article, we propose CoRe, an automated medical image processing pipeline for the prediction of postoperative LR complexity from preoperative CT scans, using imaging biomarkers. The CoRe pipeline first segments the liver, lesions, and vessels with two deep learning networks. The liver vasculature is then pruned based on a topological criterion to define the hepatic central zone (HCZ), a convex volume circumscribing the major liver vessels, from which a new imaging biomarker, BHCZ is derived. Additional biomarkers are extracted and leveraged to train and evaluate a LR complexity prediction model. An ablation study shows the HCZ-based biomarker as the central feature in predicting LR complexity. The best predictive model reaches an accuracy, F1, and AUC of 77.3, 75.4, and 84.1% respectively.
Benchmarking off-the-shelf statistical shape modeling tools in clinical applications
Sep 07, 2020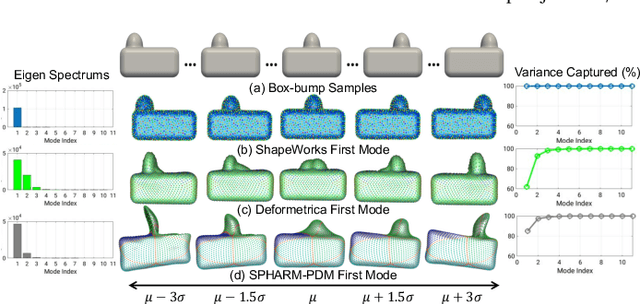

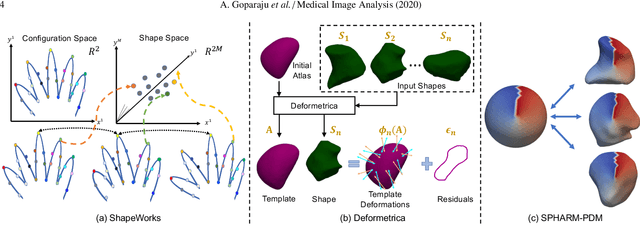
Abstract:Statistical shape modeling (SSM) is widely used in biology and medicine as a new generation of morphometric approaches for the quantitative analysis of anatomical shapes. Technological advancements of in vivo imaging have led to the development of open-source computational tools that automate the modeling of anatomical shapes and their population-level variability. However, little work has been done on the evaluation and validation of such tools in clinical applications that rely on morphometric quantifications (e.g., implant design and lesion screening). Here, we systematically assess the outcome of widely used, state-of-the-art SSM tools, namely ShapeWorks, Deformetrica, and SPHARM-PDM. We use both quantitative and qualitative metrics to evaluate shape models from different tools. We propose validation frameworks for anatomical landmark/measurement inference and lesion screening. We also present a lesion screening method to objectively characterize subtle abnormal shape changes with respect to learned population-level statistics of controls. Results demonstrate that SSM tools display different levels of consistencies, where ShapeWorks and Deformetrica models are more consistent compared to models from SPHARM-PDM due to the groupwise approach of estimating surface correspondences. Furthermore, ShapeWorks and Deformetrica shape models are found to capture clinically relevant population-level variability compared to SPHARM-PDM models.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge