Nassir Marrouche
Weighted Temporal Decay Loss for Learning Wearable PPG Data with Sparse Clinical Labels
Feb 02, 2026Abstract:Advances in wearable computing and AI have increased interest in leveraging PPG for health monitoring over the past decade. One of the biggest challenges in developing health algorithms based on such biosignals is the sparsity of clinical labels, which makes biosignals temporally distant from lab draws less reliable for supervision. To address this problem, we introduce a simple training strategy that learns a biomarker-specific decay of sample weight over the time gap between a segment and its ground truth label and uses this weight in the loss with a regularizer to prevent trivial solutions. On smartwatch PPG from 450 participants across 10 biomarkers, the approach improves over baselines. In the subject-wise setting, the proposed approach averages 0.715 AUPRC, compared to 0.674 for a fine-tuned self-supervised baseline and 0.626 for a feature-based Random Forest. A comparison of four decay families shows that a simple linear decay function is most robust on average. Beyond accuracy, the learned decay rates summarize how quickly each biomarker's PPG evidence becomes stale, providing an interpretable view of temporal sensitivity.
SOFA: Deep Learning Framework for Simulating and Optimizing Atrial Fibrillation Ablation
Aug 11, 2025



Abstract:Atrial fibrillation (AF) is a prevalent cardiac arrhythmia often treated with catheter ablation procedures, but procedural outcomes are highly variable. Evaluating and improving ablation efficacy is challenging due to the complex interaction between patient-specific tissue and procedural factors. This paper asks two questions: Can AF recurrence be predicted by simulating the effects of procedural parameters? How should we ablate to reduce AF recurrence? We propose SOFA (Simulating and Optimizing Atrial Fibrillation Ablation), a novel deep-learning framework that addresses these questions. SOFA first simulates the outcome of an ablation strategy by generating a post-ablation image depicting scar formation, conditioned on a patient's pre-ablation LGE-MRI and the specific procedural parameters used (e.g., ablation locations, duration, temperature, power, and force). During this simulation, it predicts AF recurrence risk. Critically, SOFA then introduces an optimization scheme that refines these procedural parameters to minimize the predicted risk. Our method leverages a multi-modal, multi-view generator that processes 2.5D representations of the atrium. Quantitative evaluations show that SOFA accurately synthesizes post-ablation images and that our optimization scheme leads to a 22.18\% reduction in the model-predicted recurrence risk. To the best of our knowledge, SOFA is the first framework to integrate the simulation of procedural effects, recurrence prediction, and parameter optimization, offering a novel tool for personalizing AF ablation.
SoK: Can Synthetic Images Replace Real Data? A Survey of Utility and Privacy of Synthetic Image Generation
Jun 24, 2025Abstract:Advances in generative models have transformed the field of synthetic image generation for privacy-preserving data synthesis (PPDS). However, the field lacks a comprehensive survey and comparison of synthetic image generation methods across diverse settings. In particular, when we generate synthetic images for the purpose of training a classifier, there is a pipeline of generation-sampling-classification which takes private training as input and outputs the final classifier of interest. In this survey, we systematically categorize existing image synthesis methods, privacy attacks, and mitigations along this generation-sampling-classification pipeline. To empirically compare diverse synthesis approaches, we provide a benchmark with representative generative methods and use model-agnostic membership inference attacks (MIAs) as a measure of privacy risk. Through this study, we seek to answer critical questions in PPDS: Can synthetic data effectively replace real data? Which release strategy balances utility and privacy? Do mitigations improve the utility-privacy tradeoff? Which generative models perform best across different scenarios? With a systematic evaluation of diverse methods, our study provides actionable insights into the utility-privacy tradeoffs of synthetic data generation methods and guides the decision on optimal data releasing strategies for real-world applications.
FBA-Net: Foreground and Background Aware Contrastive Learning for Semi-Supervised Atrium Segmentation
Jun 27, 2023Abstract:Medical image segmentation of gadolinium enhancement magnetic resonance imaging (GE MRI) is an important task in clinical applications. However, manual annotation is time-consuming and requires specialized expertise. Semi-supervised segmentation methods that leverage both labeled and unlabeled data have shown promise, with contrastive learning emerging as a particularly effective approach. In this paper, we propose a contrastive learning strategy of foreground and background representations for semi-supervised 3D medical image segmentation (FBA-Net). Specifically, we leverage the contrastive loss to learn representations of both the foreground and background regions in the images. By training the network to distinguish between foreground-background pairs, we aim to learn a representation that can effectively capture the anatomical structures of interest. Experiments on three medical segmentation datasets demonstrate state-of-the-art performance. Notably, our method achieves a Dice score of 91.31% with only 20% labeled data, which is remarkably close to the 91.62% score of the fully supervised method that uses 100% labeled data on the left atrium dataset. Our framework has the potential to advance the field of semi-supervised 3D medical image segmentation and enable more efficient and accurate analysis of medical images with a limited amount of annotated labels.
Benchmarking off-the-shelf statistical shape modeling tools in clinical applications
Sep 07, 2020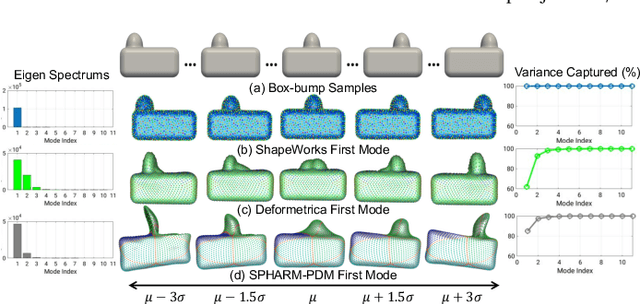

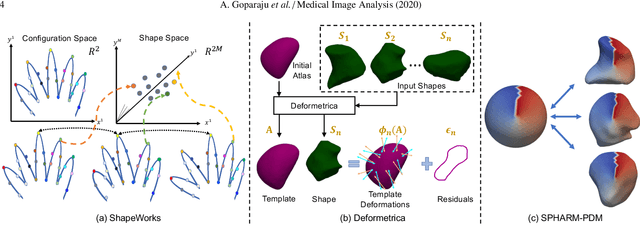
Abstract:Statistical shape modeling (SSM) is widely used in biology and medicine as a new generation of morphometric approaches for the quantitative analysis of anatomical shapes. Technological advancements of in vivo imaging have led to the development of open-source computational tools that automate the modeling of anatomical shapes and their population-level variability. However, little work has been done on the evaluation and validation of such tools in clinical applications that rely on morphometric quantifications (e.g., implant design and lesion screening). Here, we systematically assess the outcome of widely used, state-of-the-art SSM tools, namely ShapeWorks, Deformetrica, and SPHARM-PDM. We use both quantitative and qualitative metrics to evaluate shape models from different tools. We propose validation frameworks for anatomical landmark/measurement inference and lesion screening. We also present a lesion screening method to objectively characterize subtle abnormal shape changes with respect to learned population-level statistics of controls. Results demonstrate that SSM tools display different levels of consistencies, where ShapeWorks and Deformetrica models are more consistent compared to models from SPHARM-PDM due to the groupwise approach of estimating surface correspondences. Furthermore, ShapeWorks and Deformetrica shape models are found to capture clinically relevant population-level variability compared to SPHARM-PDM models.
Mixture Modeling of Global Shape Priors and Autoencoding Local Intensity Priors for Left Atrium Segmentation
Mar 06, 2019



Abstract:Difficult image segmentation problems, for instance left atrium MRI, can be addressed by incorporating shape priors to find solutions that are consistent with known objects. Nonetheless, a single multivariate Gaussian is not an adequate model in cases with significant nonlinear shape variation or where the prior distribution is multimodal. Nonparametric density estimation is more general, but has a ravenous appetite for training samples and poses serious challenges in optimization, especially in high dimensional spaces. Here, we propose a maximum-a-posteriori formulation that relies on a generative image model by incorporating both local intensity and global shape priors. We use deep autoencoders to capture the complex intensity distribution while avoiding the careful selection of hand-crafted features. We formulate the shape prior as a mixture of Gaussians and learn the corresponding parameters in a high-dimensional shape space rather than pre-projecting onto a low-dimensional subspace. In segmentation, we treat the identity of the mixture component as a latent variable and marginalize it within a generalized expectation-maximization framework. We present a conditional maximization-based scheme that alternates between a closed-form solution for component-specific shape parameters that provides a global update-based optimization strategy, and an intensity-based energy minimization that translates the global notion of a nonlinear shape prior into a set of local penalties. We demonstrate our approach on the left atrial segmentation from gadolinium-enhanced MRI, which is useful in quantifying the atrial geometry in patients with atrial fibrillation.
* Statistical Atlases and Computational Models of the Heart. Atrial Segmentation and LV Quantification Challenges 2019
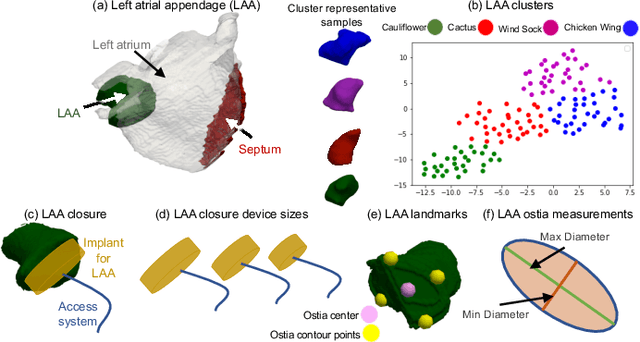
On the Evaluation and Validation of Off-the-shelf Statistical Shape Modeling Tools: A Clinical Application
Oct 03, 2018



Abstract:Statistical shape modeling (SSM) has proven useful in many areas of biology and medicine as a new generation of morphometric approaches for the quantitative analysis of anatomical shapes. Recently, the increased availability of high-resolution in vivo images of anatomy has led to the development and distribution of open-source computational tools to model anatomical shapes and their variability within populations with unprecedented detail and statistical power. Nonetheless, there is little work on the evaluation and validation of such tools as related to clinical applications that rely on morphometric quantifications for treatment planning. To address this lack of validation, we systematically assess the outcome of widely used off-the-shelf SSM tools, namely ShapeWorks, SPHARM-PDM, and Deformetrica, in the context of designing closure devices for left atrium appendage (LAA) in atrial fibrillation (AF) patients to prevent stroke, where an incomplete LAA closure may be worse than no closure. This study is motivated by the potential role of SSM in the geometric design of closure devices, which could be informed by population-level statistics, and patient-specific device selection, which is driven by anatomical measurements that could be automated by relating patient-level anatomy to population-level morphometrics. Hence, understanding the consequences of different SSM tools for the final analysis is critical for the careful choice of the tool to be deployed in real clinical scenarios. Results demonstrate that estimated measurements from ShapeWorks model are more consistent compared to models from Deformetrica and SPHARM-PDM. Furthermore, ShapeWorks and Deformetrica shape models capture clinically relevant population-level variability compared to SPHARM-PDM models.
Deep Learning for End-to-End Atrial Fibrillation Recurrence Estimation
Sep 30, 2018



Abstract:Left atrium shape has been shown to be an independent predictor of recurrence after atrial fibrillation (AF) ablation. Shape-based representation is imperative to such an estimation process, where correspondence-based representation offers the most flexibility and ease-of-computation for population-level shape statistics. Nonetheless, population-level shape representations in the form of image segmentation and correspondence models derived from cardiac MRI require significant human resources with sufficient anatomy-specific expertise. In this paper, we propose a machine learning approach that uses deep networks to estimate AF recurrence by predicting shape descriptors directly from MRI images, with NO image pre-processing involved. We also propose a novel data augmentation scheme to effectively train a deep network in a limited training data setting. We compare this new method of estimating shape descriptors from images with the state-of-the-art correspondence-based shape modeling that requires image segmentation and correspondence optimization. Results show that the proposed method and the current state-of-the-art produce statistically similar outcomes on AF recurrence, eliminating the need for expensive pre-processing pipelines and associated human labor.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge