Ross Whitaker
Audio and Multiscale Visual Cues Driven Cross-modal Transformer for Idling Vehicle Detection
Apr 15, 2025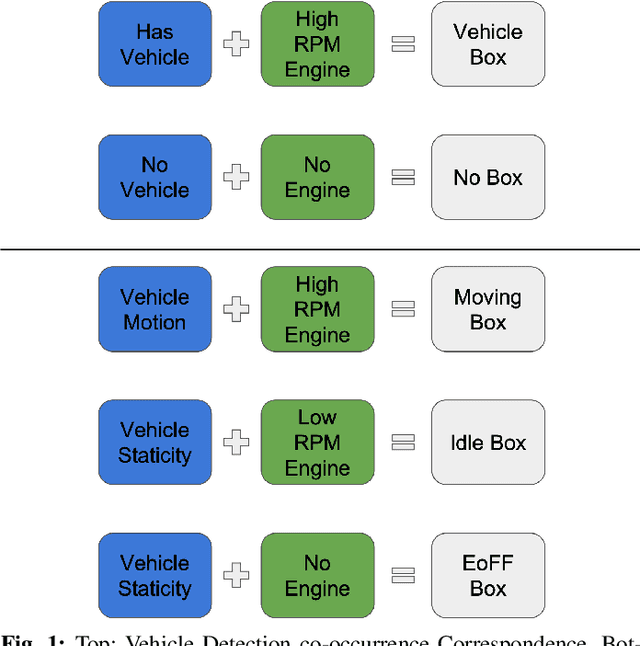
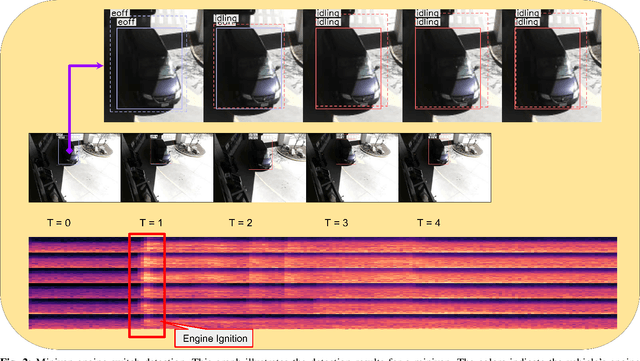
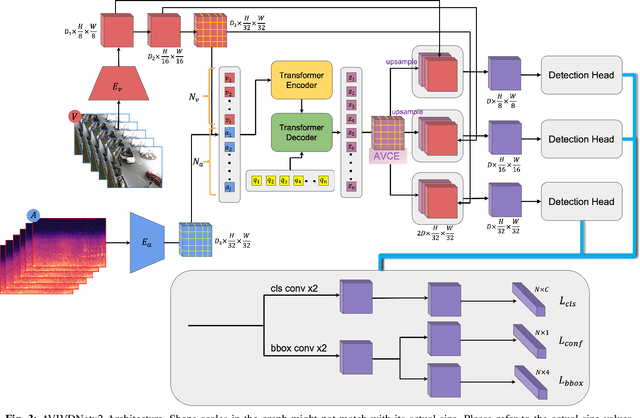
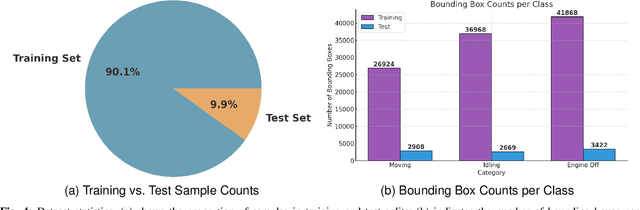
Abstract:Idling vehicle detection (IVD) supports real-time systems that reduce pollution and emissions by dynamically messaging drivers to curb excess idling behavior. In computer vision, IVD has become an emerging task that leverages video from surveillance cameras and audio from remote microphones to localize and classify vehicles in each frame as moving, idling, or engine-off. As with other cross-modal tasks, the key challenge lies in modeling the correspondence between audio and visual modalities, which differ in representation but provide complementary cues -- video offers spatial and motion context, while audio conveys engine activity beyond the visual field. The previous end-to-end model, which uses a basic attention mechanism, struggles to align these modalities effectively, often missing vehicle detections. To address this issue, we propose AVIVDNetv2, a transformer-based end-to-end detection network. It incorporates a cross-modal transformer with global patch-level learning, a multiscale visual feature fusion module, and decoupled detection heads. Extensive experiments show that AVIVDNetv2 improves mAP by 7.66 over the disjoint baseline and 9.42 over the E2E baseline, with consistent AP gains across all vehicle categories. Furthermore, AVIVDNetv2 outperforms the state-of-the-art method for sounding object localization, establishing a new performance benchmark on the AVIVD dataset.
AdaSemSeg: An Adaptive Few-shot Semantic Segmentation of Seismic Facies
Jan 28, 2025Abstract:Automated interpretation of seismic images using deep learning methods is challenging because of the limited availability of training data. Few-shot learning is a suitable learning paradigm in such scenarios due to its ability to adapt to a new task with limited supervision (small training budget). Existing few-shot semantic segmentation (FSSS) methods fix the number of target classes. Therefore, they do not support joint training on multiple datasets varying in the number of classes. In the context of the interpretation of seismic facies, fixing the number of target classes inhibits the generalization capability of a model trained on one facies dataset to another, which is likely to have a different number of facies. To address this shortcoming, we propose a few-shot semantic segmentation method for interpreting seismic facies that can adapt to the varying number of facies across the dataset, dubbed the AdaSemSeg. In general, the backbone network of FSSS methods is initialized with the statistics learned from the ImageNet dataset for better performance. The lack of such a huge annotated dataset for seismic images motivates using a self-supervised algorithm on seismic datasets to initialize the backbone network. We have trained the AdaSemSeg on three public seismic facies datasets with different numbers of facies and evaluated the proposed method on multiple metrics. The performance of the AdaSemSeg on unseen datasets (not used in training) is better than the prototype-based few-shot method and baselines.
Disentanglement Analysis in Deep Latent Variable Models Matching Aggregate Posterior Distributions
Jan 26, 2025Abstract:Deep latent variable models (DLVMs) are designed to learn meaningful representations in an unsupervised manner, such that the hidden explanatory factors are interpretable by independent latent variables (aka disentanglement). The variational autoencoder (VAE) is a popular DLVM widely studied in disentanglement analysis due to the modeling of the posterior distribution using a factorized Gaussian distribution that encourages the alignment of the latent factors with the latent axes. Several metrics have been proposed recently, assuming that the latent variables explaining the variation in data are aligned with the latent axes (cardinal directions). However, there are other DLVMs, such as the AAE and WAE-MMD (matching the aggregate posterior to the prior), where the latent variables might not be aligned with the latent axes. In this work, we propose a statistical method to evaluate disentanglement for any DLVMs in general. The proposed technique discovers the latent vectors representing the generative factors of a dataset that can be different from the cardinal latent axes. We empirically demonstrate the advantage of the method on two datasets.
ARD-VAE: A Statistical Formulation to Find the Relevant Latent Dimensions of Variational Autoencoders
Jan 18, 2025Abstract:The variational autoencoder (VAE) is a popular, deep, latent-variable model (DLVM) due to its simple yet effective formulation for modeling the data distribution. Moreover, optimizing the VAE objective function is more manageable than other DLVMs. The bottleneck dimension of the VAE is a crucial design choice, and it has strong ramifications for the model's performance, such as finding the hidden explanatory factors of a dataset using the representations learned by the VAE. However, the size of the latent dimension of the VAE is often treated as a hyperparameter estimated empirically through trial and error. To this end, we propose a statistical formulation to discover the relevant latent factors required for modeling a dataset. In this work, we use a hierarchical prior in the latent space that estimates the variance of the latent axes using the encoded data, which identifies the relevant latent dimensions. For this, we replace the fixed prior in the VAE objective function with a hierarchical prior, keeping the remainder of the formulation unchanged. We call the proposed method the automatic relevancy detection in the variational autoencoder (ARD-VAE). We demonstrate the efficacy of the ARD-VAE on multiple benchmark datasets in finding the relevant latent dimensions and their effect on different evaluation metrics, such as FID score and disentanglement analysis.
Matching aggregate posteriors in the variational autoencoder
Nov 13, 2023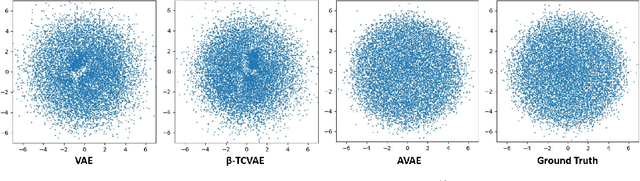

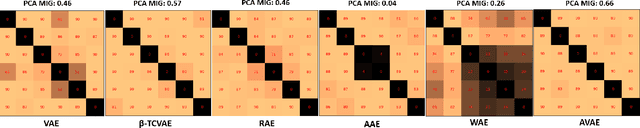
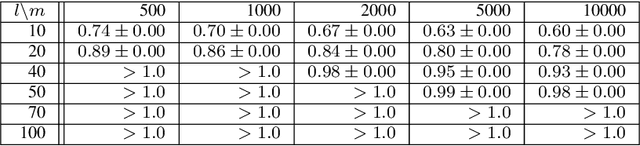
Abstract:The variational autoencoder (VAE) is a well-studied, deep, latent-variable model (DLVM) that efficiently optimizes the variational lower bound of the log marginal data likelihood and has a strong theoretical foundation. However, the VAE's known failure to match the aggregate posterior often results in \emph{pockets/holes} in the latent distribution (i.e., a failure to match the prior) and/or \emph{posterior collapse}, which is associated with a loss of information in the latent space. This paper addresses these shortcomings in VAEs by reformulating the objective function associated with VAEs in order to match the aggregate/marginal posterior distribution to the prior. We use kernel density estimate (KDE) to model the aggregate posterior in high dimensions. The proposed method is named the \emph{aggregate variational autoencoder} (AVAE) and is built on the theoretical framework of the VAE. Empirical evaluation of the proposed method on multiple benchmark data sets demonstrates the effectiveness of the AVAE relative to state-of-the-art (SOTA) methods.
Real-Time Idling Vehicles Detection Using Combined Audio-Visual Deep Learning
May 23, 2023Abstract:Combustion vehicle emissions contribute to poor air quality and release greenhouse gases into the atmosphere, and vehicle pollution has been associated with numerous adverse health effects. Roadways with extensive waiting and/or passenger drop off, such as schools and hospital drop-off zones, can result in high incidence and density of idling vehicles. This can produce micro-climates of increased vehicle pollution. Thus, the detection of idling vehicles can be helpful in monitoring and responding to unnecessary idling and be integrated into real-time or off-line systems to address the resulting pollution. In this paper we present a real-time, dynamic vehicle idling detection algorithm. The proposed idle detection algorithm and notification rely on an algorithm to detect these idling vehicles. The proposed method relies on a multi-sensor, audio-visual, machine-learning workflow to detect idling vehicles visually under three conditions: moving, static with the engine on, and static with the engine off. The visual vehicle motion detector is built in the first stage, and then a contrastive-learning-based latent space is trained for classifying static vehicle engine sound. We test our system in real-time at a hospital drop-off point in Salt Lake City. This in-situ dataset was collected and annotated, and it includes vehicles of varying models and types. The experiments show that the method can detect engine switching on or off instantly and achieves 71.01 mean average precision (mAP).
A Pathologist-Informed Workflow for Classification of Prostate Glands in Histopathology
Sep 27, 2022Abstract:Pathologists diagnose and grade prostate cancer by examining tissue from needle biopsies on glass slides. The cancer's severity and risk of metastasis are determined by the Gleason grade, a score based on the organization and morphology of prostate cancer glands. For diagnostic work-up, pathologists first locate glands in the whole biopsy core, and -- if they detect cancer -- they assign a Gleason grade. This time-consuming process is subject to errors and significant inter-observer variability, despite strict diagnostic criteria. This paper proposes an automated workflow that follows pathologists' \textit{modus operandi}, isolating and classifying multi-scale patches of individual glands in whole slide images (WSI) of biopsy tissues using distinct steps: (1) two fully convolutional networks segment epithelium versus stroma and gland boundaries, respectively; (2) a classifier network separates benign from cancer glands at high magnification; and (3) an additional classifier predicts the grade of each cancer gland at low magnification. Altogether, this process provides a gland-specific approach for prostate cancer grading that we compare against other machine-learning-based grading methods.
* Published as a workshop paper at MICCAI MOVI 2022
Leveraging Unsupervised Image Registration for Discovery of Landmark Shape Descriptor
Nov 13, 2021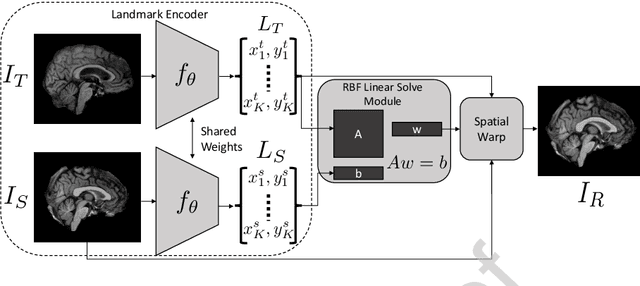
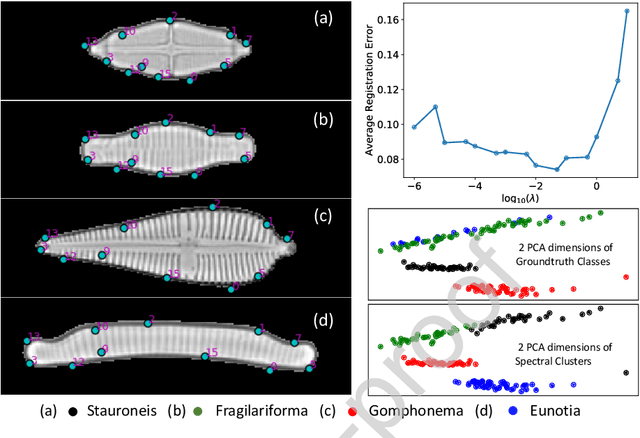
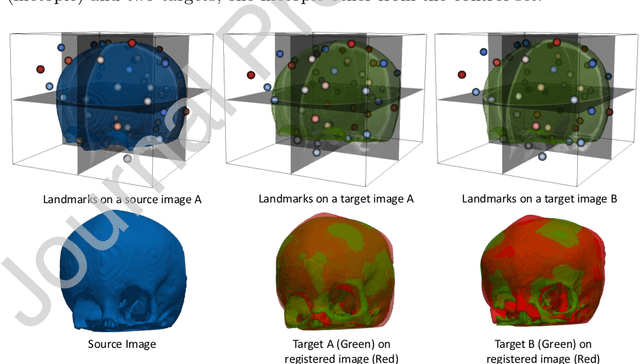
Abstract:In current biological and medical research, statistical shape modeling (SSM) provides an essential framework for the characterization of anatomy/morphology. Such analysis is often driven by the identification of a relatively small number of geometrically consistent features found across the samples of a population. These features can subsequently provide information about the population shape variation. Dense correspondence models can provide ease of computation and yield an interpretable low-dimensional shape descriptor when followed by dimensionality reduction. However, automatic methods for obtaining such correspondences usually require image segmentation followed by significant preprocessing, which is taxing in terms of both computation as well as human resources. In many cases, the segmentation and subsequent processing require manual guidance and anatomy specific domain expertise. This paper proposes a self-supervised deep learning approach for discovering landmarks from images that can directly be used as a shape descriptor for subsequent analysis. We use landmark-driven image registration as the primary task to force the neural network to discover landmarks that register the images well. We also propose a regularization term that allows for robust optimization of the neural network and ensures that the landmarks uniformly span the image domain. The proposed method circumvents segmentation and preprocessing and directly produces a usable shape descriptor using just 2D or 3D images. In addition, we also propose two variants on the training loss function that allows for prior shape information to be integrated into the model. We apply this framework on several 2D and 3D datasets to obtain their shape descriptors, and analyze their utility for various applications.
DeepSSM: A Blueprint for Image-to-Shape Deep Learning Models
Oct 14, 2021

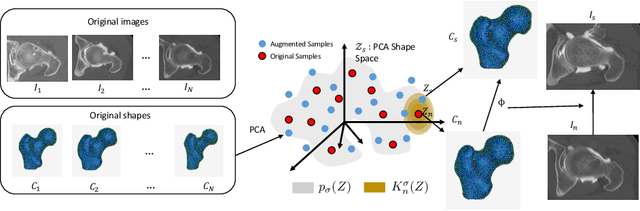
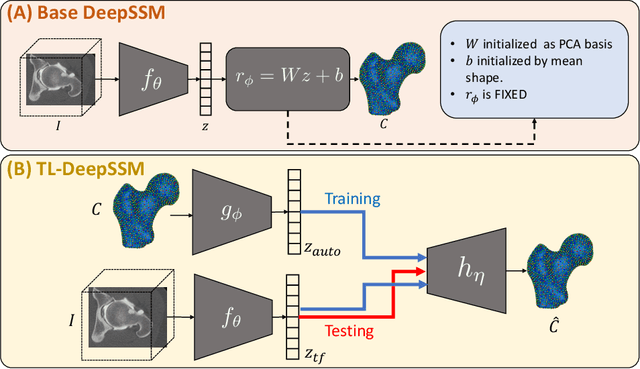
Abstract:Statistical shape modeling (SSM) characterizes anatomical variations in a population of shapes generated from medical images. SSM requires consistent shape representation across samples in shape cohort. Establishing this representation entails a processing pipeline that includes anatomy segmentation, re-sampling, registration, and non-linear optimization. These shape representations are then used to extract low-dimensional shape descriptors that facilitate subsequent analyses in different applications. However, the current process of obtaining these shape descriptors from imaging data relies on human and computational resources, requiring domain expertise for segmenting anatomies of interest. Moreover, this same taxing pipeline needs to be repeated to infer shape descriptors for new image data using a pre-trained/existing shape model. Here, we propose DeepSSM, a deep learning-based framework for learning the functional mapping from images to low-dimensional shape descriptors and their associated shape representations, thereby inferring statistical representation of anatomy directly from 3D images. Once trained using an existing shape model, DeepSSM circumvents the heavy and manual pre-processing and segmentation and significantly improves the computational time, making it a viable solution for fully end-to-end SSM applications. In addition, we introduce a model-based data-augmentation strategy to address data scarcity. Finally, this paper presents and analyzes two different architectural variants of DeepSSM with different loss functions using three medical datasets and their downstream clinical application. Experiments showcase that DeepSSM performs comparably or better to the state-of-the-art SSM both quantitatively and on application-driven downstream tasks. Therefore, DeepSSM aims to provide a comprehensive blueprint for deep learning-based image-to-shape models.
Self-Supervised Discovery of Anatomical Shape Landmarks
Jun 13, 2020
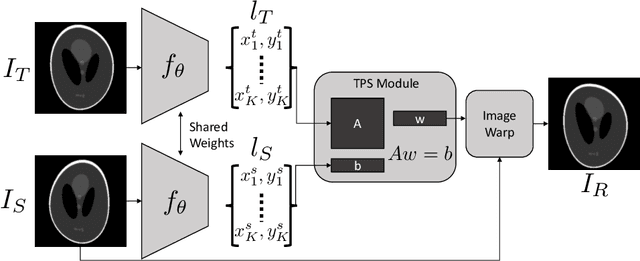


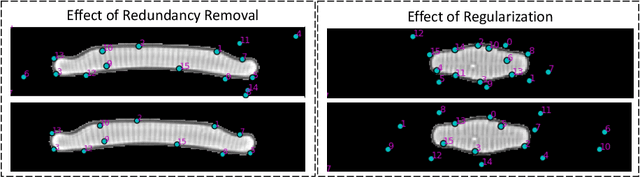
Abstract:Statistical shape analysis is a very useful tool in a wide range of medical and biological applications. However, it typically relies on the ability to produce a relatively small number of features that can capture the relevant variability in a population. State-of-the-art methods for obtaining such anatomical features rely on either extensive preprocessing or segmentation and/or significant tuning and post-processing. These shortcomings limit the widespread use of shape statistics. We propose that effective shape representations should provide sufficient information to align/register images. Using this assumption we propose a self-supervised, neural network approach for automatically positioning and detecting landmarks in images that can be used for subsequent analysis. The network discovers the landmarks corresponding to anatomical shape features that promote good image registration in the context of a particular class of transformations. In addition, we also propose a regularization for the proposed network which allows for a uniform distribution of these discovered landmarks. In this paper, we present a complete framework, which only takes a set of input images and produces landmarks that are immediately usable for statistical shape analysis. We evaluate the performance on a phantom dataset as well as 2D and 3D images.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge