Shireen Elhabian
Mesh2SSM++: A Probabilistic Framework for Unsupervised Learning of Statistical Shape Model of Anatomies from Surface Meshes
Feb 11, 2025
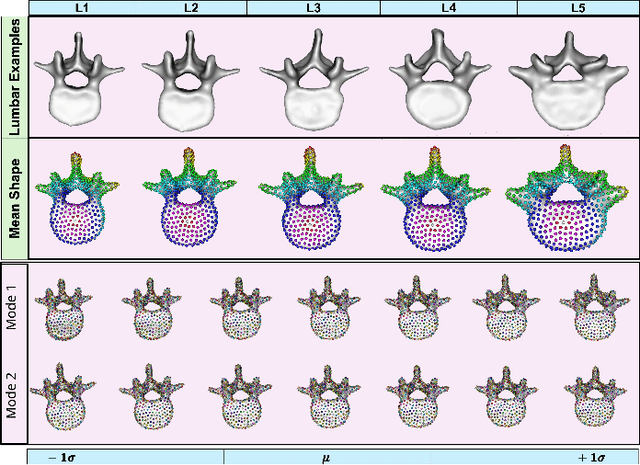
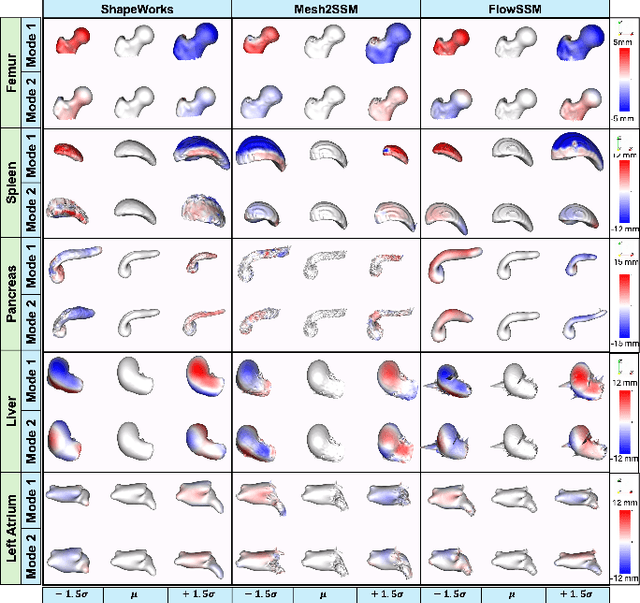
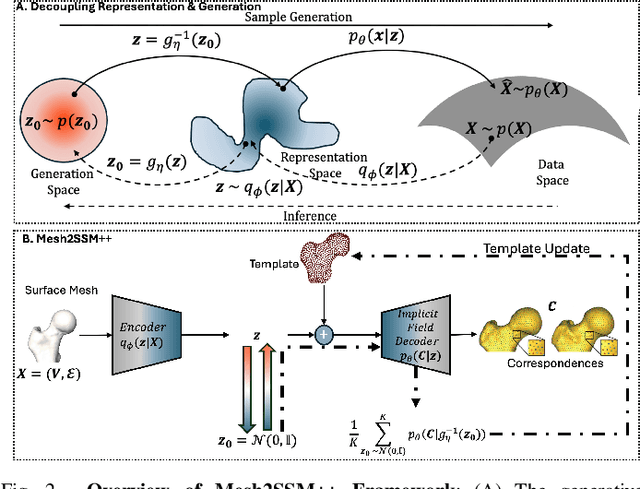
Abstract:Anatomy evaluation is crucial for understanding the physiological state, diagnosing abnormalities, and guiding medical interventions. Statistical shape modeling (SSM) is vital in this process. By enabling the extraction of quantitative morphological shape descriptors from MRI and CT scans, SSM provides comprehensive descriptions of anatomical variations within a population. However, the effectiveness of SSM in anatomy evaluation hinges on the quality and robustness of the shape models. While deep learning techniques show promise in addressing these challenges by learning complex nonlinear representations of shapes, existing models still have limitations and often require pre-established shape models for training. To overcome these issues, we propose Mesh2SSM++, a novel approach that learns to estimate correspondences from meshes in an unsupervised manner. This method leverages unsupervised, permutation-invariant representation learning to estimate how to deform a template point cloud into subject-specific meshes, forming a correspondence-based shape model. Additionally, our probabilistic formulation allows learning a population-specific template, reducing potential biases associated with template selection. A key feature of Mesh2SSM++ is its ability to quantify aleatoric uncertainty, which captures inherent data variability and is essential for ensuring reliable model predictions and robust decision-making in clinical tasks, especially under challenging imaging conditions. Through extensive validation across diverse anatomies, evaluation metrics, and downstream tasks, we demonstrate that Mesh2SSM++ outperforms existing methods. Its ability to operate directly on meshes, combined with computational efficiency and interpretability through its probabilistic framework, makes it an attractive alternative to traditional and deep learning-based SSM approaches.
MORPH-LER: Log-Euclidean Regularization for Population-Aware Image Registration
Feb 04, 2025



Abstract:Spatial transformations that capture population-level morphological statistics are critical for medical image analysis. Commonly used smoothness regularizers for image registration fail to integrate population statistics, leading to anatomically inconsistent transformations. Inverse consistency regularizers promote geometric consistency but lack population morphometrics integration. Regularizers that constrain deformation to low-dimensional manifold methods address this. However, they prioritize reconstruction over interpretability and neglect diffeomorphic properties, such as group composition and inverse consistency. We introduce MORPH-LER, a Log-Euclidean regularization framework for population-aware unsupervised image registration. MORPH-LER learns population morphometrics from spatial transformations to guide and regularize registration networks, ensuring anatomically plausible deformations. It features a bottleneck autoencoder that computes the principal logarithm of deformation fields via iterative square-root predictions. It creates a linearized latent space that respects diffeomorphic properties and enforces inverse consistency. By integrating a registration network with a diffeomorphic autoencoder, MORPH-LER produces smooth, meaningful deformation fields. The framework offers two main contributions: (1) a data-driven regularization strategy that incorporates population-level anatomical statistics to enhance transformation validity and (2) a linearized latent space that enables compact and interpretable deformation fields for efficient population morphometrics analysis. We validate MORPH-LER across two families of deep learning-based registration networks, demonstrating its ability to produce anatomically accurate, computationally efficient, and statistically meaningful transformations on the OASIS-1 brain imaging dataset.
LEDA: Log-Euclidean Diffeomorphic Autoencoder for Efficient Statistical Analysis of Diffeomorphism
Dec 20, 2024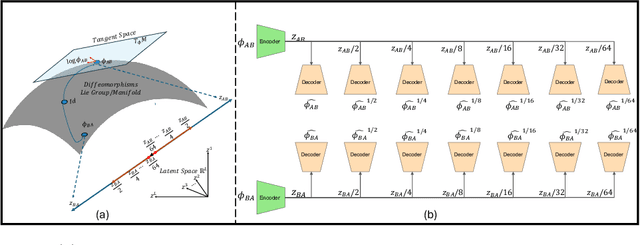
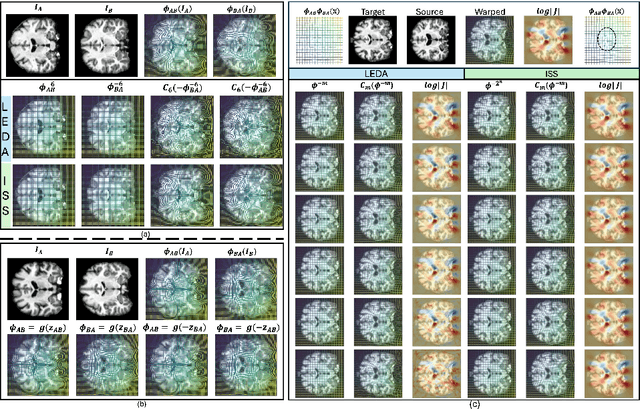
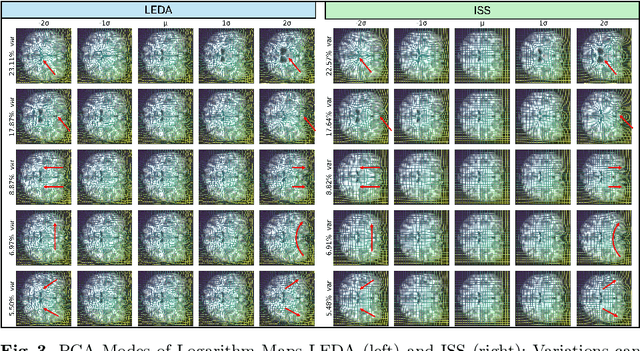
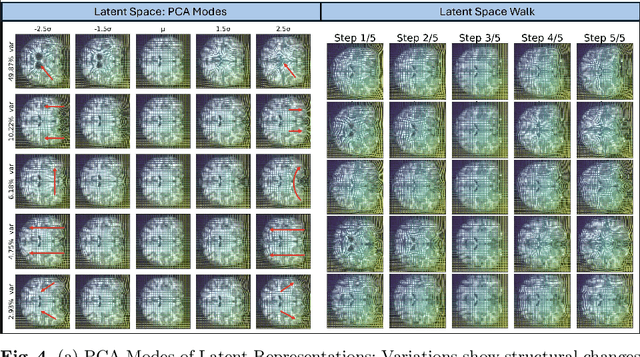
Abstract:Image registration is a core task in computational anatomy that establishes correspondences between images. Invertible deformable registration, which computes a deformation field and handles complex, non-linear transformation, is essential for tracking anatomical variations, especially in neuroimaging applications where inter-subject differences and longitudinal changes are key. Analyzing the deformation fields is challenging due to their non-linearity, limiting statistical analysis. However, traditional approaches for analyzing deformation fields are computationally expensive, sensitive to initialization, and prone to numerical errors, especially when the deformation is far from the identity. To address these limitations, we propose the Log-Euclidean Diffeomorphic Autoencoder (LEDA), an innovative framework designed to compute the principal logarithm of deformation fields by efficiently predicting consecutive square roots. LEDA operates within a linearized latent space that adheres to the diffeomorphisms group action laws, enhancing our model's robustness and applicability. We also introduce a loss function to enforce inverse consistency, ensuring accurate latent representations of deformation fields. Extensive experiments with the OASIS-1 dataset demonstrate the effectiveness of LEDA in accurately modeling and analyzing complex non-linear deformations while maintaining inverse consistency. Additionally, we evaluate its ability to capture and incorporate clinical variables, enhancing its relevance for clinical applications.
HAMIL-QA: Hierarchical Approach to Multiple Instance Learning for Atrial LGE MRI Quality Assessment
Jul 09, 2024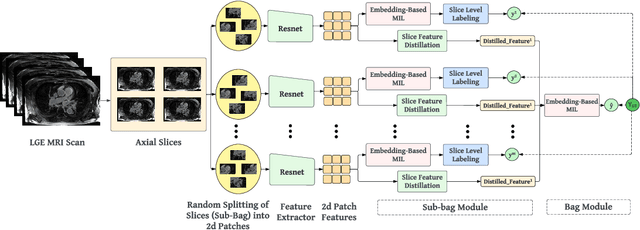
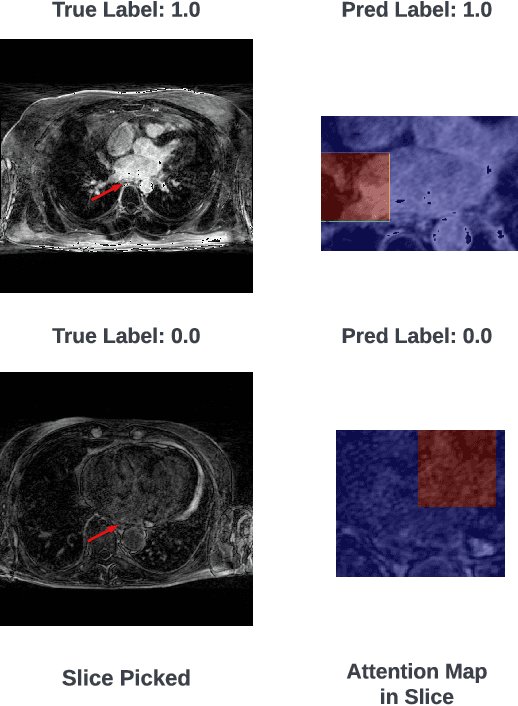
Abstract:The accurate evaluation of left atrial fibrosis via high-quality 3D Late Gadolinium Enhancement (LGE) MRI is crucial for atrial fibrillation management but is hindered by factors like patient movement and imaging variability. The pursuit of automated LGE MRI quality assessment is critical for enhancing diagnostic accuracy, standardizing evaluations, and improving patient outcomes. The deep learning models aimed at automating this process face significant challenges due to the scarcity of expert annotations, high computational costs, and the need to capture subtle diagnostic details in highly variable images. This study introduces HAMIL-QA, a multiple instance learning (MIL) framework, designed to overcome these obstacles. HAMIL-QA employs a hierarchical bag and sub-bag structure that allows for targeted analysis within sub-bags and aggregates insights at the volume level. This hierarchical MIL approach reduces reliance on extensive annotations, lessens computational load, and ensures clinically relevant quality predictions by focusing on diagnostically critical image features. Our experiments show that HAMIL-QA surpasses existing MIL methods and traditional supervised approaches in accuracy, AUROC, and F1-Score on an LGE MRI scan dataset, demonstrating its potential as a scalable solution for LGE MRI quality assessment automation. The code is available at: $\href{https://github.com/arf111/HAMIL-QA}{\text{this https URL}}$
Weakly Supervised Bayesian Shape Modeling from Unsegmented Medical Images
May 15, 2024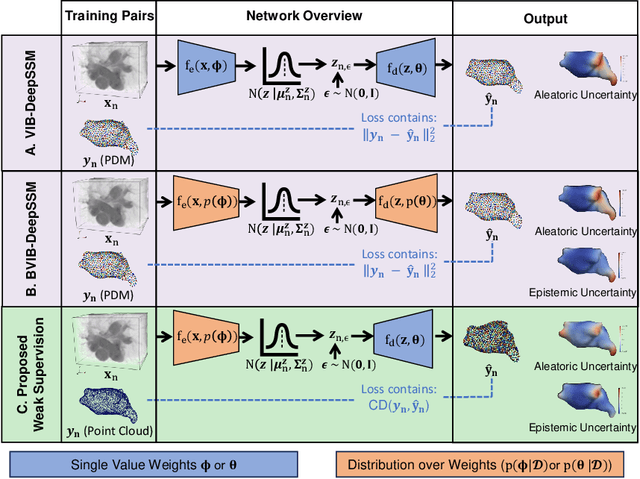
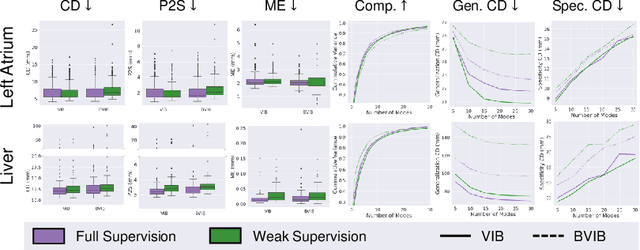
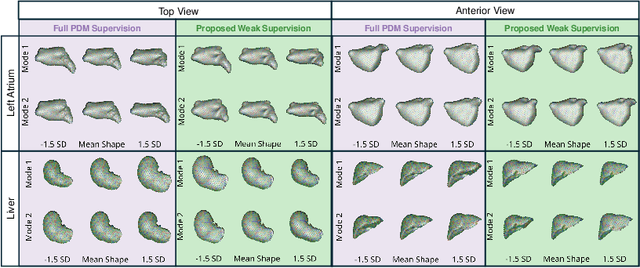
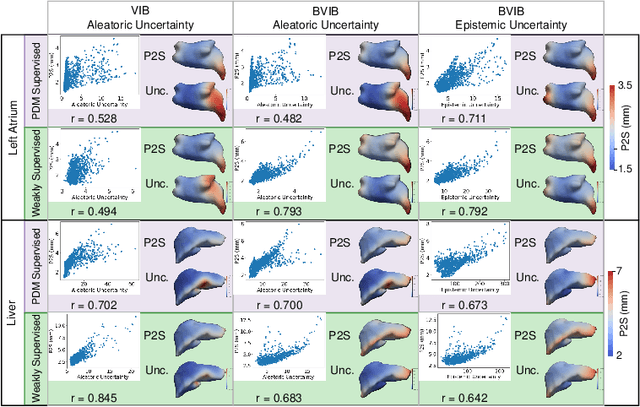
Abstract:Anatomical shape analysis plays a pivotal role in clinical research and hypothesis testing, where the relationship between form and function is paramount. Correspondence-based statistical shape modeling (SSM) facilitates population-level morphometrics but requires a cumbersome, potentially bias-inducing construction pipeline. Recent advancements in deep learning have streamlined this process in inference by providing SSM prediction directly from unsegmented medical images. However, the proposed approaches are fully supervised and require utilizing a traditional SSM construction pipeline to create training data, thus inheriting the associated burdens and limitations. To address these challenges, we introduce a weakly supervised deep learning approach to predict SSM from images using point cloud supervision. Specifically, we propose reducing the supervision associated with the state-of-the-art fully Bayesian variational information bottleneck DeepSSM (BVIB-DeepSSM) model. BVIB-DeepSSM is an effective, principled framework for predicting probabilistic anatomical shapes from images with quantification of both aleatoric and epistemic uncertainties. Whereas the original BVIB-DeepSSM method requires strong supervision in the form of ground truth correspondence points, the proposed approach utilizes weak supervision via point cloud surface representations, which are more readily obtainable. Furthermore, the proposed approach learns correspondence in a completely data-driven manner without prior assumptions about the expected variability in shape cohort. Our experiments demonstrate that this approach yields similar accuracy and uncertainty estimation to the fully supervised scenario while substantially enhancing the feasibility of model training for SSM construction.
Point2SSM++: Self-Supervised Learning of Anatomical Shape Models from Point Clouds
May 15, 2024



Abstract:Correspondence-based statistical shape modeling (SSM) stands as a powerful technology for morphometric analysis in clinical research. SSM facilitates population-level characterization and quantification of anatomical shapes such as bones and organs, aiding in pathology and disease diagnostics and treatment planning. Despite its potential, SSM remains under-utilized in medical research due to the significant overhead associated with automatic construction methods, which demand complete, aligned shape surface representations. Additionally, optimization-based techniques rely on bias-inducing assumptions or templates and have prolonged inference times as the entire cohort is simultaneously optimized. To overcome these challenges, we introduce Point2SSM++, a principled, self-supervised deep learning approach that directly learns correspondence points from point cloud representations of anatomical shapes. Point2SSM++ is robust to misaligned and inconsistent input, providing SSM that accurately samples individual shape surfaces while effectively capturing population-level statistics. Additionally, we present principled extensions of Point2SSM++ to adapt it for dynamic spatiotemporal and multi-anatomy use cases, demonstrating the broad versatility of the Point2SSM++ framework. Furthermore, we present extensions of Point2SSM++ tailored for dynamic spatiotemporal and multi-anatomy scenarios, showcasing the broad versatility of the framework. Through extensive validation across diverse anatomies, evaluation metrics, and clinically relevant downstream tasks, we demonstrate Point2SSM++'s superiority over existing state-of-the-art deep learning models and traditional approaches. Point2SSM++ substantially enhances the feasibility of SSM generation and significantly broadens its array of potential clinical applications.
Two-Stage Deep Learning Framework for Quality Assessment of Left Atrial Late Gadolinium Enhanced MRI Images
Oct 13, 2023Abstract:Accurate assessment of left atrial fibrosis in patients with atrial fibrillation relies on high-quality 3D late gadolinium enhancement (LGE) MRI images. However, obtaining such images is challenging due to patient motion, changing breathing patterns, or sub-optimal choice of pulse sequence parameters. Automated assessment of LGE-MRI image diagnostic quality is clinically significant as it would enhance diagnostic accuracy, improve efficiency, ensure standardization, and contributes to better patient outcomes by providing reliable and high-quality LGE-MRI scans for fibrosis quantification and treatment planning. To address this, we propose a two-stage deep-learning approach for automated LGE-MRI image diagnostic quality assessment. The method includes a left atrium detector to focus on relevant regions and a deep network to evaluate diagnostic quality. We explore two training strategies, multi-task learning, and pretraining using contrastive learning, to overcome limited annotated data in medical imaging. Contrastive Learning result shows about $4\%$, and $9\%$ improvement in F1-Score and Specificity compared to Multi-Task learning when there's limited data.
Progressive DeepSSM: Training Methodology for Image-To-Shape Deep Models
Oct 02, 2023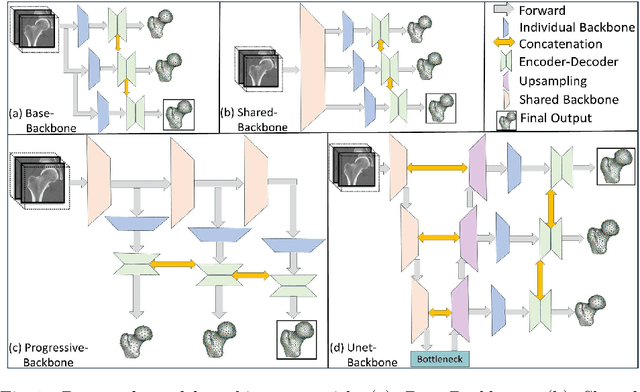
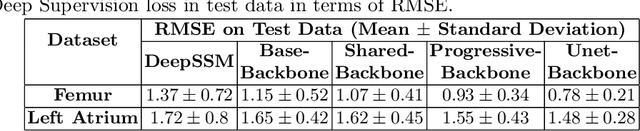
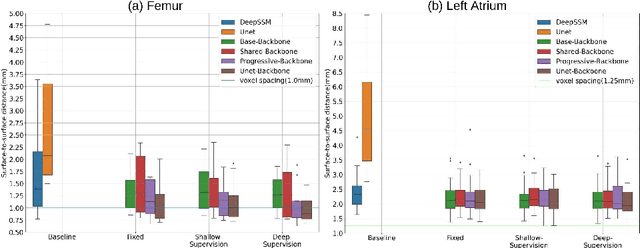

Abstract:Statistical shape modeling (SSM) is an enabling quantitative tool to study anatomical shapes in various medical applications. However, directly using 3D images in these applications still has a long way to go. Recent deep learning methods have paved the way for reducing the substantial preprocessing steps to construct SSMs directly from unsegmented images. Nevertheless, the performance of these models is not up to the mark. Inspired by multiscale/multiresolution learning, we propose a new training strategy, progressive DeepSSM, to train image-to-shape deep learning models. The training is performed in multiple scales, and each scale utilizes the output from the previous scale. This strategy enables the model to learn coarse shape features in the first scales and gradually learn detailed fine shape features in the later scales. We leverage shape priors via segmentation-guided multi-task learning and employ deep supervision loss to ensure learning at each scale. Experiments show the superiority of models trained by the proposed strategy from both quantitative and qualitative perspectives. This training methodology can be employed to improve the stability and accuracy of any deep learning method for inferring statistical representations of anatomies from medical images and can be adopted by existing deep learning methods to improve model accuracy and training stability.
MedShapeNet -- A Large-Scale Dataset of 3D Medical Shapes for Computer Vision
Sep 12, 2023



Abstract:We present MedShapeNet, a large collection of anatomical shapes (e.g., bones, organs, vessels) and 3D surgical instrument models. Prior to the deep learning era, the broad application of statistical shape models (SSMs) in medical image analysis is evidence that shapes have been commonly used to describe medical data. Nowadays, however, state-of-the-art (SOTA) deep learning algorithms in medical imaging are predominantly voxel-based. In computer vision, on the contrary, shapes (including, voxel occupancy grids, meshes, point clouds and implicit surface models) are preferred data representations in 3D, as seen from the numerous shape-related publications in premier vision conferences, such as the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR), as well as the increasing popularity of ShapeNet (about 51,300 models) and Princeton ModelNet (127,915 models) in computer vision research. MedShapeNet is created as an alternative to these commonly used shape benchmarks to facilitate the translation of data-driven vision algorithms to medical applications, and it extends the opportunities to adapt SOTA vision algorithms to solve critical medical problems. Besides, the majority of the medical shapes in MedShapeNet are modeled directly on the imaging data of real patients, and therefore it complements well existing shape benchmarks comprising of computer-aided design (CAD) models. MedShapeNet currently includes more than 100,000 medical shapes, and provides annotations in the form of paired data. It is therefore also a freely available repository of 3D models for extended reality (virtual reality - VR, augmented reality - AR, mixed reality - MR) and medical 3D printing. This white paper describes in detail the motivations behind MedShapeNet, the shape acquisition procedures, the use cases, as well as the usage of the online shape search portal: https://medshapenet.ikim.nrw/
ADASSM: Adversarial Data Augmentation in Statistical Shape Models From Images
Jul 10, 2023



Abstract:Statistical shape models (SSM) have been well-established as an excellent tool for identifying variations in the morphology of anatomy across the underlying population. Shape models use consistent shape representation across all the samples in a given cohort, which helps to compare shapes and identify the variations that can detect pathologies and help in formulating treatment plans. In medical imaging, computing these shape representations from CT/MRI scans requires time-intensive preprocessing operations, including but not limited to anatomy segmentation annotations, registration, and texture denoising. Deep learning models have demonstrated exceptional capabilities in learning shape representations directly from volumetric images, giving rise to highly effective and efficient Image-to-SSM. Nevertheless, these models are data-hungry and due to the limited availability of medical data, deep learning models tend to overfit. Offline data augmentation techniques, that use kernel density estimation based (KDE) methods for generating shape-augmented samples, have successfully aided Image-to-SSM networks in achieving comparable accuracy to traditional SSM methods. However, these augmentation methods focus on shape augmentation, whereas deep learning models exhibit image-based texture bias results in sub-optimal models. This paper introduces a novel strategy for on-the-fly data augmentation for the Image-to-SSM framework by leveraging data-dependent noise generation or texture augmentation. The proposed framework is trained as an adversary to the Image-to-SSM network, augmenting diverse and challenging noisy samples. Our approach achieves improved accuracy by encouraging the model to focus on the underlying geometry rather than relying solely on pixel values.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge