Sarang Joshi
Scientific Computing and Imaging Institute, University of Utah, Department of Biomedical Engineering, University of Utah
Domain-Shift Immunity in Deep Deformable Registration via Local Feature Representations
Dec 29, 2025Abstract:Deep learning has advanced deformable image registration, surpassing traditional optimization-based methods in both accuracy and efficiency. However, learning-based models are widely believed to be sensitive to domain shift, with robustness typically pursued through large and diverse training datasets, without explaining the underlying mechanisms. In this work, we show that domain-shift immunity is an inherent property of deep deformable registration models, arising from their reliance on local feature representations rather than global appearance for deformation estimation. To isolate and validate this mechanism, we introduce UniReg, a universal registration framework that decouples feature extraction from deformation estimation using fixed, pre-trained feature extractors and a UNet-based deformation network. Despite training on a single dataset, UniReg exhibits robust cross-domain and multi-modal performance comparable to optimization-based methods. Our analysis further reveals that failures of conventional CNN-based models under modality shift originate from dataset-induced biases in early convolutional layers. These findings identify local feature consistency as the key driver of robustness in learning-based deformable registration and motivate backbone designs that preserve domain-invariant local features.
Tackling Hallucination from Conditional Models for Medical Image Reconstruction with DynamicDPS
Mar 03, 2025Abstract:Hallucinations are spurious structures not present in the ground truth, posing a critical challenge in medical image reconstruction, especially for data-driven conditional models. We hypothesize that combining an unconditional diffusion model with data consistency, trained on a diverse dataset, can reduce these hallucinations. Based on this, we propose DynamicDPS, a diffusion-based framework that integrates conditional and unconditional diffusion models to enhance low-quality medical images while systematically reducing hallucinations. Our approach first generates an initial reconstruction using a conditional model, then refines it with an adaptive diffusion-based inverse problem solver. DynamicDPS skips early stage in the reverse process by selecting an optimal starting time point per sample and applies Wolfe's line search for adaptive step sizes, improving both efficiency and image fidelity. Using diffusion priors and data consistency, our method effectively reduces hallucinations from any conditional model output. We validate its effectiveness in Image Quality Transfer for low-field MRI enhancement. Extensive evaluations on synthetic and real MR scans, including a downstream task for tissue volume estimation, show that DynamicDPS reduces hallucinations, improving relative volume estimation by over 15% for critical tissues while using only 5% of the sampling steps required by baseline diffusion models. As a model-agnostic and fine-tuning-free approach, DynamicDPS offers a robust solution for hallucination reduction in medical imaging. The code will be made publicly available upon publication.
MORPH-LER: Log-Euclidean Regularization for Population-Aware Image Registration
Feb 04, 2025



Abstract:Spatial transformations that capture population-level morphological statistics are critical for medical image analysis. Commonly used smoothness regularizers for image registration fail to integrate population statistics, leading to anatomically inconsistent transformations. Inverse consistency regularizers promote geometric consistency but lack population morphometrics integration. Regularizers that constrain deformation to low-dimensional manifold methods address this. However, they prioritize reconstruction over interpretability and neglect diffeomorphic properties, such as group composition and inverse consistency. We introduce MORPH-LER, a Log-Euclidean regularization framework for population-aware unsupervised image registration. MORPH-LER learns population morphometrics from spatial transformations to guide and regularize registration networks, ensuring anatomically plausible deformations. It features a bottleneck autoencoder that computes the principal logarithm of deformation fields via iterative square-root predictions. It creates a linearized latent space that respects diffeomorphic properties and enforces inverse consistency. By integrating a registration network with a diffeomorphic autoencoder, MORPH-LER produces smooth, meaningful deformation fields. The framework offers two main contributions: (1) a data-driven regularization strategy that incorporates population-level anatomical statistics to enhance transformation validity and (2) a linearized latent space that enables compact and interpretable deformation fields for efficient population morphometrics analysis. We validate MORPH-LER across two families of deep learning-based registration networks, demonstrating its ability to produce anatomically accurate, computationally efficient, and statistically meaningful transformations on the OASIS-1 brain imaging dataset.
Disentanglement Analysis in Deep Latent Variable Models Matching Aggregate Posterior Distributions
Jan 26, 2025Abstract:Deep latent variable models (DLVMs) are designed to learn meaningful representations in an unsupervised manner, such that the hidden explanatory factors are interpretable by independent latent variables (aka disentanglement). The variational autoencoder (VAE) is a popular DLVM widely studied in disentanglement analysis due to the modeling of the posterior distribution using a factorized Gaussian distribution that encourages the alignment of the latent factors with the latent axes. Several metrics have been proposed recently, assuming that the latent variables explaining the variation in data are aligned with the latent axes (cardinal directions). However, there are other DLVMs, such as the AAE and WAE-MMD (matching the aggregate posterior to the prior), where the latent variables might not be aligned with the latent axes. In this work, we propose a statistical method to evaluate disentanglement for any DLVMs in general. The proposed technique discovers the latent vectors representing the generative factors of a dataset that can be different from the cardinal latent axes. We empirically demonstrate the advantage of the method on two datasets.
ARD-VAE: A Statistical Formulation to Find the Relevant Latent Dimensions of Variational Autoencoders
Jan 18, 2025Abstract:The variational autoencoder (VAE) is a popular, deep, latent-variable model (DLVM) due to its simple yet effective formulation for modeling the data distribution. Moreover, optimizing the VAE objective function is more manageable than other DLVMs. The bottleneck dimension of the VAE is a crucial design choice, and it has strong ramifications for the model's performance, such as finding the hidden explanatory factors of a dataset using the representations learned by the VAE. However, the size of the latent dimension of the VAE is often treated as a hyperparameter estimated empirically through trial and error. To this end, we propose a statistical formulation to discover the relevant latent factors required for modeling a dataset. In this work, we use a hierarchical prior in the latent space that estimates the variance of the latent axes using the encoded data, which identifies the relevant latent dimensions. For this, we replace the fixed prior in the VAE objective function with a hierarchical prior, keeping the remainder of the formulation unchanged. We call the proposed method the automatic relevancy detection in the variational autoencoder (ARD-VAE). We demonstrate the efficacy of the ARD-VAE on multiple benchmark datasets in finding the relevant latent dimensions and their effect on different evaluation metrics, such as FID score and disentanglement analysis.
The Silent Majority: Demystifying Memorization Effect in the Presence of Spurious Correlations
Jan 01, 2025Abstract:Machine learning models often rely on simple spurious features -- patterns in training data that correlate with targets but are not causally related to them, like image backgrounds in foreground classification. This reliance typically leads to imbalanced test performance across minority and majority groups. In this work, we take a closer look at the fundamental cause of such imbalanced performance through the lens of memorization, which refers to the ability to predict accurately on \textit{atypical} examples (minority groups) in the training set but failing in achieving the same accuracy in the testing set. This paper systematically shows the ubiquitous existence of spurious features in a small set of neurons within the network, providing the first-ever evidence that memorization may contribute to imbalanced group performance. Through three experimental sources of converging empirical evidence, we find the property of a small subset of neurons or channels in memorizing minority group information. Inspired by these findings, we articulate the hypothesis: the imbalanced group performance is a byproduct of ``noisy'' spurious memorization confined to a small set of neurons. To further substantiate this hypothesis, we show that eliminating these unnecessary spurious memorization patterns via a novel framework during training can significantly affect the model performance on minority groups. Our experimental results across various architectures and benchmarks offer new insights on how neural networks encode core and spurious knowledge, laying the groundwork for future research in demystifying robustness to spurious correlation.
LEDA: Log-Euclidean Diffeomorphic Autoencoder for Efficient Statistical Analysis of Diffeomorphism
Dec 20, 2024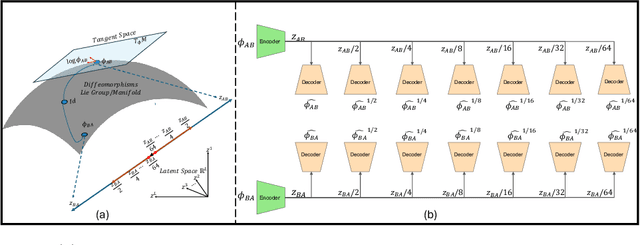
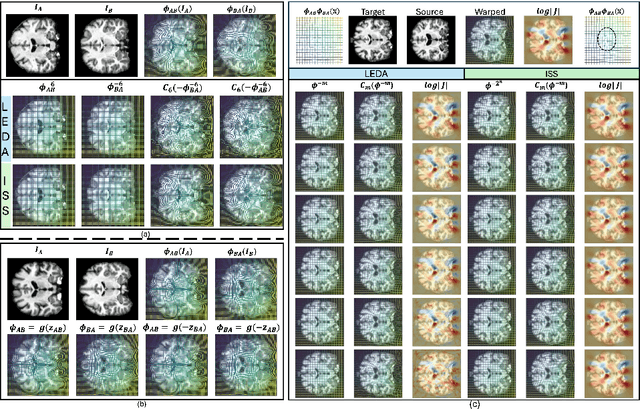
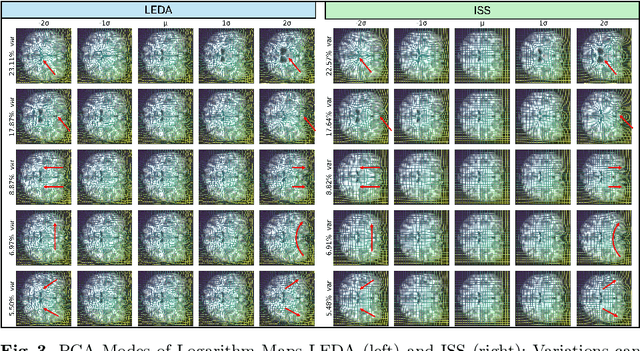
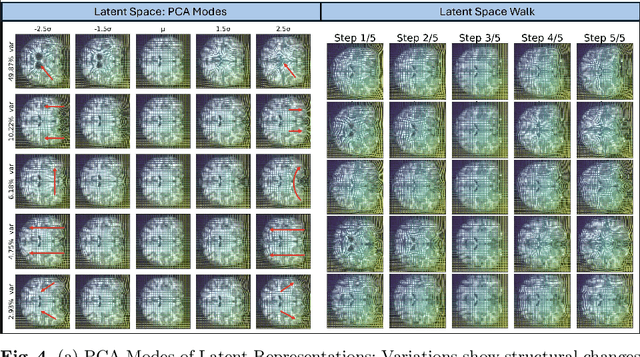
Abstract:Image registration is a core task in computational anatomy that establishes correspondences between images. Invertible deformable registration, which computes a deformation field and handles complex, non-linear transformation, is essential for tracking anatomical variations, especially in neuroimaging applications where inter-subject differences and longitudinal changes are key. Analyzing the deformation fields is challenging due to their non-linearity, limiting statistical analysis. However, traditional approaches for analyzing deformation fields are computationally expensive, sensitive to initialization, and prone to numerical errors, especially when the deformation is far from the identity. To address these limitations, we propose the Log-Euclidean Diffeomorphic Autoencoder (LEDA), an innovative framework designed to compute the principal logarithm of deformation fields by efficiently predicting consecutive square roots. LEDA operates within a linearized latent space that adheres to the diffeomorphisms group action laws, enhancing our model's robustness and applicability. We also introduce a loss function to enforce inverse consistency, ensuring accurate latent representations of deformation fields. Extensive experiments with the OASIS-1 dataset demonstrate the effectiveness of LEDA in accurately modeling and analyzing complex non-linear deformations while maintaining inverse consistency. Additionally, we evaluate its ability to capture and incorporate clinical variables, enhancing its relevance for clinical applications.
Refining Skewed Perceptions in Vision-Language Models through Visual Representations
May 22, 2024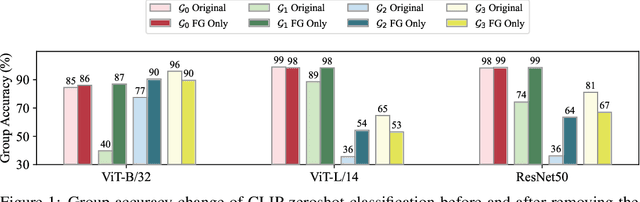

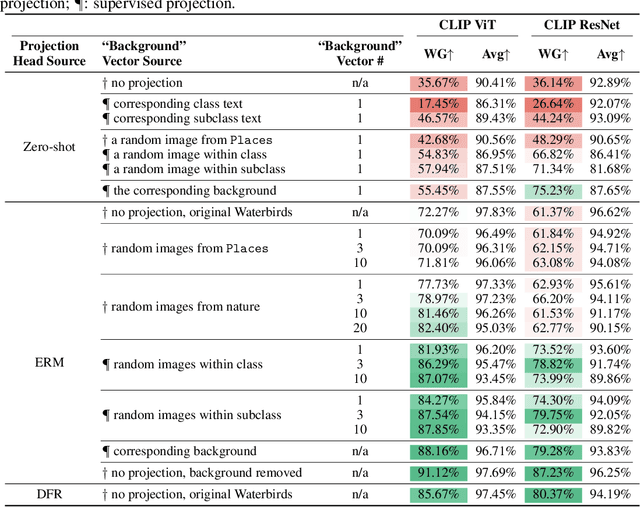
Abstract:Large vision-language models (VLMs), such as CLIP, have become foundational, demonstrating remarkable success across a variety of downstream tasks. Despite their advantages, these models, akin to other foundational systems, inherit biases from the disproportionate distribution of real-world data, leading to misconceptions about the actual environment. Prevalent datasets like ImageNet are often riddled with non-causal, spurious correlations that can diminish VLM performance in scenarios where these contextual elements are absent. This study presents an investigation into how a simple linear probe can effectively distill task-specific core features from CLIP's embedding for downstream applications. Our analysis reveals that the CLIP text representations are often tainted by spurious correlations, inherited in the biased pre-training dataset. Empirical evidence suggests that relying on visual representations from CLIP, as opposed to text embedding, is more practical to refine the skewed perceptions in VLMs, emphasizing the superior utility of visual representations in overcoming embedded biases. Our codes will be available here.
Matching aggregate posteriors in the variational autoencoder
Nov 13, 2023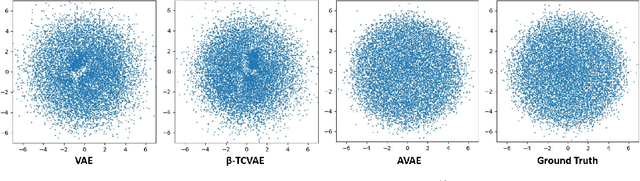

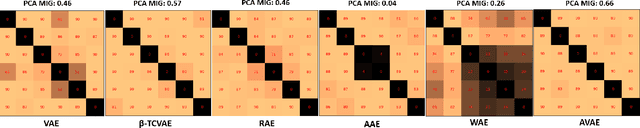
Abstract:The variational autoencoder (VAE) is a well-studied, deep, latent-variable model (DLVM) that efficiently optimizes the variational lower bound of the log marginal data likelihood and has a strong theoretical foundation. However, the VAE's known failure to match the aggregate posterior often results in \emph{pockets/holes} in the latent distribution (i.e., a failure to match the prior) and/or \emph{posterior collapse}, which is associated with a loss of information in the latent space. This paper addresses these shortcomings in VAEs by reformulating the objective function associated with VAEs in order to match the aggregate/marginal posterior distribution to the prior. We use kernel density estimate (KDE) to model the aggregate posterior in high dimensions. The proposed method is named the \emph{aggregate variational autoencoder} (AVAE) and is built on the theoretical framework of the VAE. Empirical evaluation of the proposed method on multiple benchmark data sets demonstrates the effectiveness of the AVAE relative to state-of-the-art (SOTA) methods.
Analyzing the Domain Shift Immunity of Deep Homography Estimation
Apr 19, 2023Abstract:Homography estimation is a basic image-alignment method in many applications. Recently, with the development of convolutional neural networks (CNNs), some learning based approaches have shown great success in this task. However, the performance across different domains has never been researched. Unlike other common tasks (\eg, classification, detection, segmentation), CNN based homography estimation models show a domain shift immunity, which means a model can be trained on one dataset and tested on another without any transfer learning. To explain this unusual performance, we need to determine how CNNs estimate homography. In this study, we first show the domain shift immunity of different deep homography estimation models. We then use a shallow network with a specially designed dataset to analyze the features used for estimation. The results show that networks use low-level texture information to estimate homography. We also design some experiments to compare the performance between different texture densities and image features distorted on some common datasets to demonstrate our findings. Based on these findings, we provide an explanation of the domain shift immunity of deep homography estimation.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge