Tolga Tasdizen
How to Build Robust, Scalable Models for GSV-Based Indicators in Neighborhood Research
Jan 10, 2026Abstract:A substantial body of health research demonstrates a strong link between neighborhood environments and health outcomes. Recently, there has been increasing interest in leveraging advances in computer vision to enable large-scale, systematic characterization of neighborhood built environments. However, the generalizability of vision models across fundamentally different domains remains uncertain, for example, transferring knowledge from ImageNet to the distinct visual characteristics of Google Street View (GSV) imagery. In applied fields such as social health research, several critical questions arise: which models are most appropriate, whether to adopt unsupervised training strategies, what training scale is feasible under computational constraints, and how much such strategies benefit downstream performance. These decisions are often costly and require specialized expertise. In this paper, we answer these questions through empirical analysis and provide practical insights into how to select and adapt foundation models for datasets with limited size and labels, while leveraging larger, unlabeled datasets through unsupervised training. Our study includes comprehensive quantitative and visual analyses comparing model performance before and after unsupervised adaptation.
DuoFormer: Leveraging Hierarchical Representations by Local and Global Attention Vision Transformer
Jun 15, 2025Abstract:Despite the widespread adoption of transformers in medical applications, the exploration of multi-scale learning through transformers remains limited, while hierarchical representations are considered advantageous for computer-aided medical diagnosis. We propose a novel hierarchical transformer model that adeptly integrates the feature extraction capabilities of Convolutional Neural Networks (CNNs) with the advanced representational potential of Vision Transformers (ViTs). Addressing the lack of inductive biases and dependence on extensive training datasets in ViTs, our model employs a CNN backbone to generate hierarchical visual representations. These representations are adapted for transformer input through an innovative patch tokenization process, preserving the inherited multi-scale inductive biases. We also introduce a scale-wise attention mechanism that directly captures intra-scale and inter-scale associations. This mechanism complements patch-wise attention by enhancing spatial understanding and preserving global perception, which we refer to as local and global attention, respectively. Our model significantly outperforms baseline models in terms of classification accuracy, demonstrating its efficiency in bridging the gap between Convolutional Neural Networks (CNNs) and Vision Transformers (ViTs). The components are designed as plug-and-play for different CNN architectures and can be adapted for multiple applications. The code is available at https://github.com/xiaoyatang/DuoFormer.git.
Audio and Multiscale Visual Cues Driven Cross-modal Transformer for Idling Vehicle Detection
Apr 15, 2025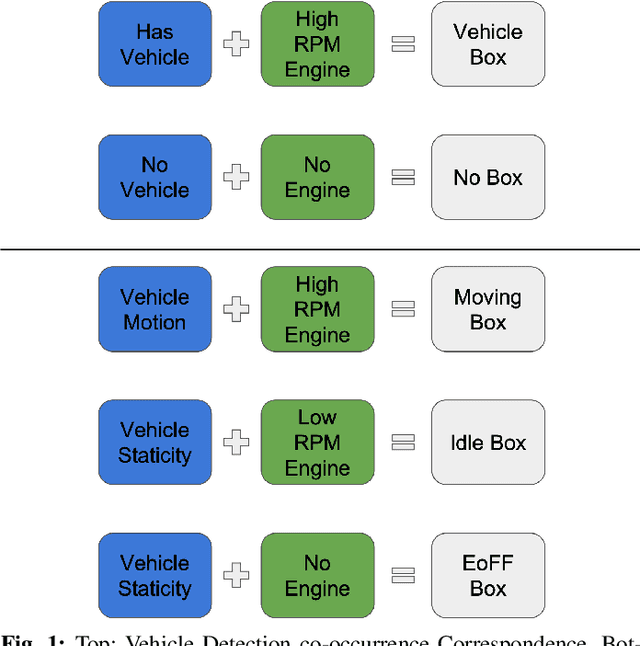
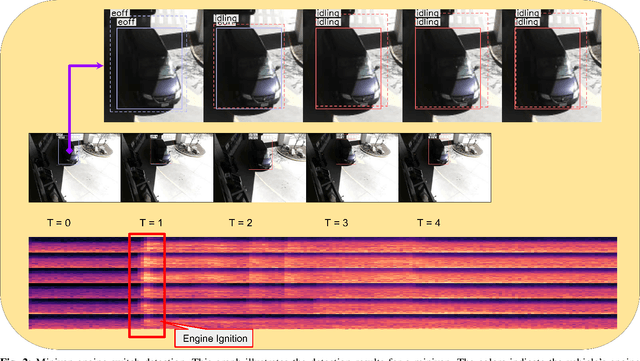
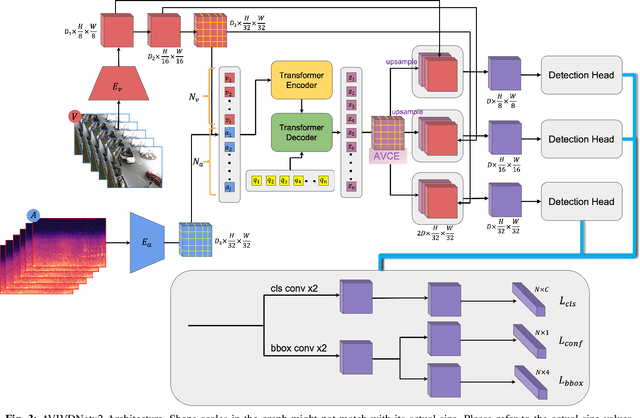
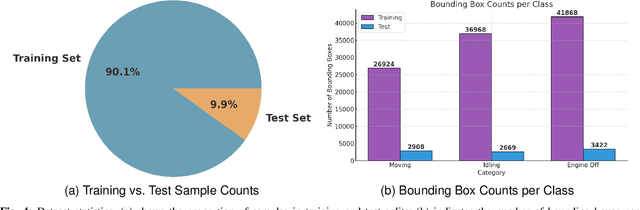
Abstract:Idling vehicle detection (IVD) supports real-time systems that reduce pollution and emissions by dynamically messaging drivers to curb excess idling behavior. In computer vision, IVD has become an emerging task that leverages video from surveillance cameras and audio from remote microphones to localize and classify vehicles in each frame as moving, idling, or engine-off. As with other cross-modal tasks, the key challenge lies in modeling the correspondence between audio and visual modalities, which differ in representation but provide complementary cues -- video offers spatial and motion context, while audio conveys engine activity beyond the visual field. The previous end-to-end model, which uses a basic attention mechanism, struggles to align these modalities effectively, often missing vehicle detections. To address this issue, we propose AVIVDNetv2, a transformer-based end-to-end detection network. It incorporates a cross-modal transformer with global patch-level learning, a multiscale visual feature fusion module, and decoupled detection heads. Extensive experiments show that AVIVDNetv2 improves mAP by 7.66 over the disjoint baseline and 9.42 over the E2E baseline, with consistent AP gains across all vehicle categories. Furthermore, AVIVDNetv2 outperforms the state-of-the-art method for sounding object localization, establishing a new performance benchmark on the AVIVD dataset.
WeakSupCon: Weakly Supervised Contrastive Learning for Encoder Pre-training
Mar 06, 2025



Abstract:Weakly supervised multiple instance learning (MIL) is a challenging task given that only bag-level labels are provided, while each bag typically contains multiple instances. This topic has been extensively studied in histopathological image analysis, where labels are usually available only at the whole slide image (WSI) level, while each whole slide image can be divided into thousands of small image patches for training. The dominant MIL approaches take fixed patch features as inputs to address computational constraints and ensure model stability. These features are commonly generated by encoders pre-trained on ImageNet, foundation encoders pre-trained on large datasets, or through self-supervised learning on local datasets. While the self-supervised encoder pre-training on the same dataset as downstream MIL tasks helps mitigate domain shift and generate better features, the bag-level labels are not utilized during the process, and the features of patches from different categories may cluster together, reducing classification performance on MIL tasks. Recently, pre-training with supervised contrastive learning (SupCon) has demonstrated superior performance compared to self-supervised contrastive learning and even end-to-end training on traditional image classification tasks. In this paper, we propose a novel encoder pre-training method for downstream MIL tasks called Weakly Supervised Contrastive Learning (WeakSupCon) that utilizes bag-level labels. In our method, we employ multi-task learning and define distinct contrastive learning losses for samples with different bag labels. Our experiments demonstrate that the features generated using WeakSupCon significantly enhance MIL classification performance compared to self-supervised approaches across three datasets.
A Comparison of Object Detection and Phrase Grounding Models in Chest X-ray Abnormality Localization using Eye-tracking Data
Mar 02, 2025Abstract:Chest diseases rank among the most prevalent and dangerous global health issues. Object detection and phrase grounding deep learning models interpret complex radiology data to assist healthcare professionals in diagnosis. Object detection locates abnormalities for classes, while phrase grounding locates abnormalities for textual descriptions. This paper investigates how text enhances abnormality localization in chest X-rays by comparing the performance and explainability of these two tasks. To establish an explainability baseline, we proposed an automatic pipeline to generate image regions for report sentences using radiologists' eye-tracking data. The better performance - mIoU = 0.36 vs. 0.20 - and explainability - Containment ratio 0.48 vs. 0.26 - of the phrase grounding model infers the effectiveness of text in enhancing chest X-ray abnormality localization.
Hierarchical Transformer for Electrocardiogram Diagnosis
Nov 01, 2024Abstract:Transformers, originally prominent in NLP and computer vision, are now being adapted for ECG signal analysis. This paper introduces a novel hierarchical transformer architecture that segments the model into multiple stages by assessing the spatial size of the embeddings, thus eliminating the need for additional downsampling strategies or complex attention designs. A classification token aggregates information across feature scales, facilitating interactions between different stages of the transformer. By utilizing depth-wise convolutions in a six-layer convolutional encoder, our approach preserves the relationships between different ECG leads. Moreover, an attention gate mechanism learns associations among the leads prior to classification. This model adapts flexibly to various embedding networks and input sizes while enhancing the interpretability of transformers in ECG signal analysis.
SRA: A Novel Method to Improve Feature Embedding in Self-supervised Learning for Histopathological Images
Oct 31, 2024



Abstract:Self-supervised learning has become a cornerstone in various areas, particularly histopathological image analysis. Image augmentation plays a crucial role in self-supervised learning, as it generates variations in image samples. However, traditional image augmentation techniques often overlook the unique characteristics of histopathological images. In this paper, we propose a new histopathology-specific image augmentation method called stain reconstruction augmentation (SRA). We integrate our SRA with MoCo v3, a leading model in self-supervised contrastive learning, along with our additional contrastive loss terms, and call the new model SRA-MoCo v3. We demonstrate that our SRA-MoCo v3 always outperforms the standard MoCo v3 across various downstream tasks and achieves comparable or superior performance to other foundation models pre-trained on significantly larger histopathology datasets.
Joint Audio-Visual Idling Vehicle Detection with Streamlined Input Dependencies
Oct 28, 2024

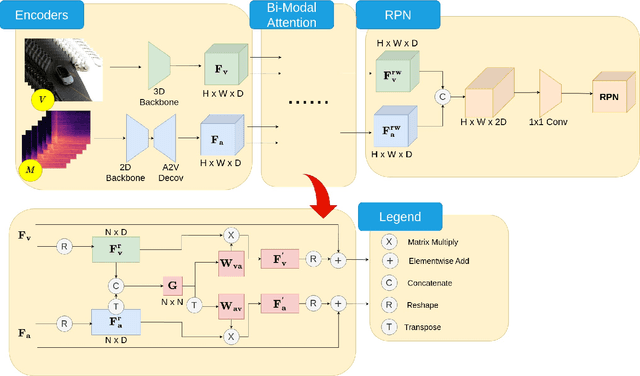

Abstract:Idling vehicle detection (IVD) can be helpful in monitoring and reducing unnecessary idling and can be integrated into real-time systems to address the resulting pollution and harmful products. The previous approach [13], a non-end-to-end model, requires extra user clicks to specify a part of the input, making system deployment more error-prone or even not feasible. In contrast, we introduce an end-to-end joint audio-visual IVD task designed to detect vehicles visually under three states: moving, idling and engine off. Unlike feature co-occurrence task such as audio-visual vehicle tracking, our IVD task addresses complementary features, where labels cannot be determined by a single modality alone. To this end, we propose AVIVD-Net, a novel network that integrates audio and visual features through a bidirectional attention mechanism. AVIVD-Net streamlines the input process by learning a joint feature space, reducing the deployment complexity of previous methods. Additionally, we introduce the AVIVD dataset, which is seven times larger than previous datasets, offering significantly more annotated samples to study the IVD problem. Our model achieves performance comparable to prior approaches, making it suitable for automated deployment. Furthermore, by evaluating AVIVDNet on the feature co-occurrence public dataset MAVD [23], we demonstrate its potential for extension to self-driving vehicle video-camera setups.
PathMoCo: A Novel Framework to Improve Feature Embedding in Self-supervised Contrastive Learning for Histopathological Images
Oct 23, 2024



Abstract:Self-supervised contrastive learning has become a cornerstone in various areas, particularly histopathological image analysis. Image augmentation plays a crucial role in self-supervised contrastive learning, as it generates variations in image samples. However, traditional image augmentation techniques often overlook the unique characteristics of histopathological images. In this paper, we propose a new histopathology-specific image augmentation method called stain reconstruction augmentation (SRA). We integrate our SRA with MoCo v3, a leading model in self-supervised contrastive learning, along with our additional contrastive loss terms, and call the new model PathMoCo. We demonstrate that our PathMoCo always outperforms the standard MoCo v3 across various downstream tasks and achieves comparable or superior performance to other foundation models pre-trained on significantly larger histopathology datasets.
VISTA: A Visual and Textual Attention Dataset for Interpreting Multimodal Models
Oct 06, 2024Abstract:The recent developments in deep learning led to the integration of natural language processing (NLP) with computer vision, resulting in powerful integrated Vision and Language Models (VLMs). Despite their remarkable capabilities, these models are frequently regarded as black boxes within the machine learning research community. This raises a critical question: which parts of an image correspond to specific segments of text, and how can we decipher these associations? Understanding these connections is essential for enhancing model transparency, interpretability, and trustworthiness. To answer this question, we present an image-text aligned human visual attention dataset that maps specific associations between image regions and corresponding text segments. We then compare the internal heatmaps generated by VL models with this dataset, allowing us to analyze and better understand the model's decision-making process. This approach aims to enhance model transparency, interpretability, and trustworthiness by providing insights into how these models align visual and linguistic information. We conducted a comprehensive study on text-guided visual saliency detection in these VL models. This study aims to understand how different models prioritize and focus on specific visual elements in response to corresponding text segments, providing deeper insights into their internal mechanisms and improving our ability to interpret their outputs.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge