Yosep Chong
VIMs: Virtual Immunohistochemistry Multiplex staining via Text-to-Stain Diffusion Trained on Uniplex Stains
Jul 26, 2024
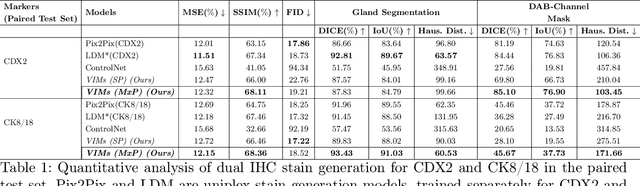

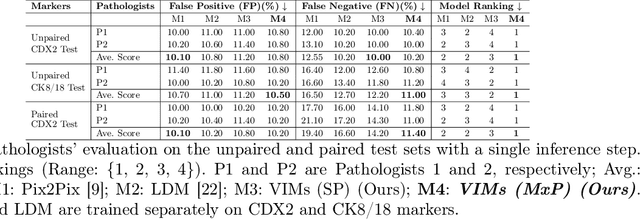
Abstract:This paper introduces a Virtual Immunohistochemistry Multiplex staining (VIMs) model designed to generate multiple immunohistochemistry (IHC) stains from a single hematoxylin and eosin (H&E) stained tissue section. IHC stains are crucial in pathology practice for resolving complex diagnostic questions and guiding patient treatment decisions. While commercial laboratories offer a wide array of up to 400 different antibody-based IHC stains, small biopsies often lack sufficient tissue for multiple stains while preserving material for subsequent molecular testing. This highlights the need for virtual IHC staining. Notably, VIMs is the first model to address this need, leveraging a large vision-language single-step diffusion model for virtual IHC multiplexing through text prompts for each IHC marker. VIMs is trained on uniplex paired H&E and IHC images, employing an adversarial training module. Testing of VIMs includes both paired and unpaired image sets. To enhance computational efficiency, VIMs utilizes a pre-trained large latent diffusion model fine-tuned with small, trainable weights through the Low-Rank Adapter (LoRA) approach. Experiments on nuclear and cytoplasmic IHC markers demonstrate that VIMs outperforms the base diffusion model and achieves performance comparable to Pix2Pix, a standard generative model for paired image translation. Multiple evaluation methods, including assessments by two pathologists, are used to determine the performance of VIMs. Additionally, experiments with different prompts highlight the impact of text conditioning. This paper represents the first attempt to accelerate histopathology research by demonstrating the generation of multiple IHC stains from a single H&E input using a single model trained solely on uniplex data.
DISC: Latent Diffusion Models with Self-Distillation from Separated Conditions for Prostate Cancer Grading
Apr 19, 2024Abstract:Latent Diffusion Models (LDMs) can generate high-fidelity images from noise, offering a promising approach for augmenting histopathology images for training cancer grading models. While previous works successfully generated high-fidelity histopathology images using LDMs, the generation of image tiles to improve prostate cancer grading has not yet been explored. Additionally, LDMs face challenges in accurately generating admixtures of multiple cancer grades in a tile when conditioned by a tile mask. In this study, we train specific LDMs to generate synthetic tiles that contain multiple Gleason Grades (GGs) by leveraging pixel-wise annotations in input tiles. We introduce a novel framework named Self-Distillation from Separated Conditions (DISC) that generates GG patterns guided by GG masks. Finally, we deploy a training framework for pixel-level and slide-level prostate cancer grading, where synthetic tiles are effectively utilized to improve the cancer grading performance of existing models. As a result, this work surpasses previous works in two domains: 1) our LDMs enhanced with DISC produce more accurate tiles in terms of GG patterns, and 2) our training scheme, incorporating synthetic data, significantly improves the generalization of the baseline model for prostate cancer grading, particularly in challenging cases of rare GG5, demonstrating the potential of generative models to enhance cancer grading when data is limited.
F2FLDM: Latent Diffusion Models with Histopathology Pre-Trained Embeddings for Unpaired Frozen Section to FFPE Translation
Apr 19, 2024



Abstract:The Frozen Section (FS) technique is a rapid and efficient method, taking only 15-30 minutes to prepare slides for pathologists' evaluation during surgery, enabling immediate decisions on further surgical interventions. However, FS process often introduces artifacts and distortions like folds and ice-crystal effects. In contrast, these artifacts and distortions are absent in the higher-quality formalin-fixed paraffin-embedded (FFPE) slides, which require 2-3 days to prepare. While Generative Adversarial Network (GAN)-based methods have been used to translate FS to FFPE images (F2F), they may leave morphological inaccuracies with remaining FS artifacts or introduce new artifacts, reducing the quality of these translations for clinical assessments. In this study, we benchmark recent generative models, focusing on GANs and Latent Diffusion Models (LDMs), to overcome these limitations. We introduce a novel approach that combines LDMs with Histopathology Pre-Trained Embeddings to enhance restoration of FS images. Our framework leverages LDMs conditioned by both text and pre-trained embeddings to learn meaningful features of FS and FFPE histopathology images. Through diffusion and denoising techniques, our approach not only preserves essential diagnostic attributes like color staining and tissue morphology but also proposes an embedding translation mechanism to better predict the targeted FFPE representation of input FS images. As a result, this work achieves a significant improvement in classification performance, with the Area Under the Curve rising from 81.99% to 94.64%, accompanied by an advantageous CaseFD. This work establishes a new benchmark for FS to FFPE image translation quality, promising enhanced reliability and accuracy in histopathology FS image analysis. Our work is available at https://minhmanho.github.io/f2f_ldm/.
CLASS-M: Adaptive stain separation-based contrastive learning with pseudo-labeling for histopathological image classification
Jan 04, 2024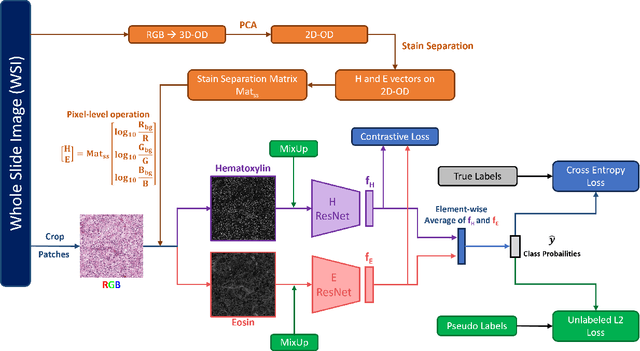
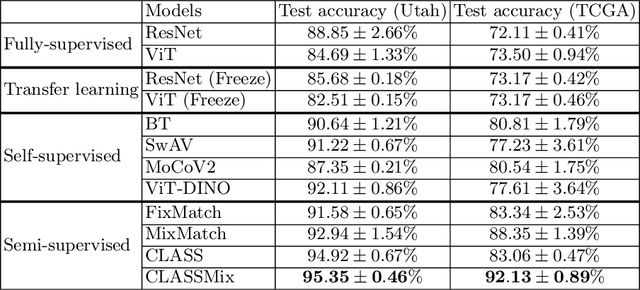
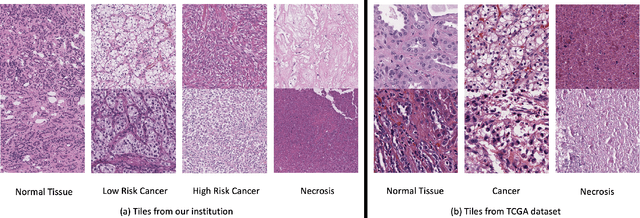
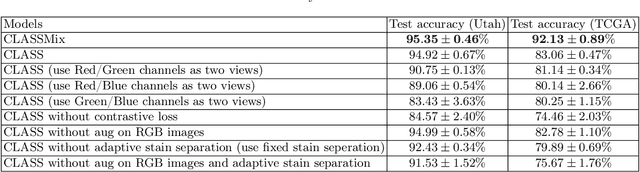
Abstract:Histopathological image classification is an important task in medical image analysis. Recent approaches generally rely on weakly supervised learning due to the ease of acquiring case-level labels from pathology reports. However, patch-level classification is preferable in applications where only a limited number of cases are available or when local prediction accuracy is critical. On the other hand, acquiring extensive datasets with localized labels for training is not feasible. In this paper, we propose a semi-supervised patch-level histopathological image classification model, named CLASS-M, that does not require extensively labeled datasets. CLASS-M is formed by two main parts: a contrastive learning module that uses separated Hematoxylin and Eosin images generated through an adaptive stain separation process, and a module with pseudo-labels using MixUp. We compare our model with other state-of-the-art models on two clear cell renal cell carcinoma datasets. We demonstrate that our CLASS-M model has the best performance on both datasets. Our code is available at github.com/BzhangURU/Paper_CLASS-M/tree/main
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge