VIMs: Virtual Immunohistochemistry Multiplex staining via Text-to-Stain Diffusion Trained on Uniplex Stains
Paper and Code
Jul 26, 2024
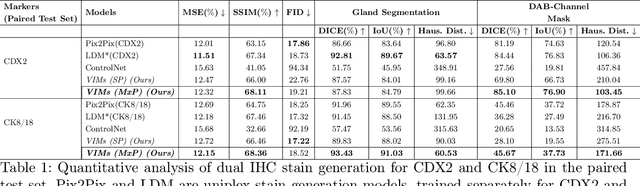

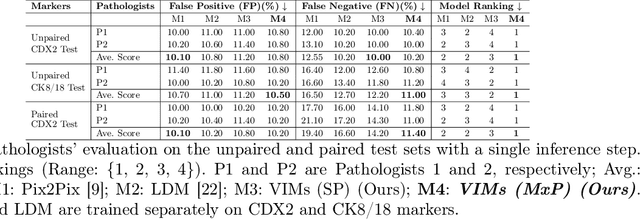
This paper introduces a Virtual Immunohistochemistry Multiplex staining (VIMs) model designed to generate multiple immunohistochemistry (IHC) stains from a single hematoxylin and eosin (H&E) stained tissue section. IHC stains are crucial in pathology practice for resolving complex diagnostic questions and guiding patient treatment decisions. While commercial laboratories offer a wide array of up to 400 different antibody-based IHC stains, small biopsies often lack sufficient tissue for multiple stains while preserving material for subsequent molecular testing. This highlights the need for virtual IHC staining. Notably, VIMs is the first model to address this need, leveraging a large vision-language single-step diffusion model for virtual IHC multiplexing through text prompts for each IHC marker. VIMs is trained on uniplex paired H&E and IHC images, employing an adversarial training module. Testing of VIMs includes both paired and unpaired image sets. To enhance computational efficiency, VIMs utilizes a pre-trained large latent diffusion model fine-tuned with small, trainable weights through the Low-Rank Adapter (LoRA) approach. Experiments on nuclear and cytoplasmic IHC markers demonstrate that VIMs outperforms the base diffusion model and achieves performance comparable to Pix2Pix, a standard generative model for paired image translation. Multiple evaluation methods, including assessments by two pathologists, are used to determine the performance of VIMs. Additionally, experiments with different prompts highlight the impact of text conditioning. This paper represents the first attempt to accelerate histopathology research by demonstrating the generation of multiple IHC stains from a single H&E input using a single model trained solely on uniplex data.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge