Shireen Y. Elhabian
Building Trust in Virtual Immunohistochemistry: Automated Assessment of Image Quality
Nov 06, 2025



Abstract:Deep learning models can generate virtual immunohistochemistry (IHC) stains from hematoxylin and eosin (H&E) images, offering a scalable and low-cost alternative to laboratory IHC. However, reliable evaluation of image quality remains a challenge as current texture- and distribution-based metrics quantify image fidelity rather than the accuracy of IHC staining. Here, we introduce an automated and accuracy grounded framework to determine image quality across sixteen paired or unpaired image translation models. Using color deconvolution, we generate masks of pixels stained brown (i.e., IHC-positive) as predicted by each virtual IHC model. We use the segmented masks of real and virtual IHC to compute stain accuracy metrics (Dice, IoU, Hausdorff distance) that directly quantify correct pixel - level labeling without needing expert manual annotations. Our results demonstrate that conventional image fidelity metrics, including Frechet Inception Distance (FID), peak signal-to-noise ratio (PSNR), and structural similarity (SSIM), correlate poorly with stain accuracy and pathologist assessment. Paired models such as PyramidPix2Pix and AdaptiveNCE achieve the highest stain accuracy, whereas unpaired diffusion- and GAN-based models are less reliable in providing accurate IHC positive pixel labels. Moreover, whole-slide images (WSI) reveal performance declines that are invisible in patch-based evaluations, emphasizing the need for WSI-level benchmarks. Together, this framework defines a reproducible approach for assessing the quality of virtual IHC models, a critical step to accelerate translation towards routine use by pathologists.
Adaptive Particle-Based Shape Modeling for Anatomical Surface Correspondence
Jul 10, 2025Abstract:Particle-based shape modeling (PSM) is a family of approaches that automatically quantifies shape variability across anatomical cohorts by positioning particles (pseudo landmarks) on shape surfaces in a consistent configuration. Recent advances incorporate implicit radial basis function representations as self-supervised signals to better capture the complex geometric properties of anatomical structures. However, these methods still lack self-adaptivity -- that is, the ability to automatically adjust particle configurations to local geometric features of each surface, which is essential for accurately representing complex anatomical variability. This paper introduces two mechanisms to increase surface adaptivity while maintaining consistent particle configurations: (1) a novel neighborhood correspondence loss to enable high adaptivity and (2) a geodesic correspondence algorithm that regularizes optimization to enforce geodesic neighborhood consistency. We evaluate the efficacy and scalability of our approach on challenging datasets, providing a detailed analysis of the adaptivity-correspondence trade-off and benchmarking against existing methods on surface representation accuracy and correspondence metrics.
ImplicitStainer: Data-Efficient Medical Image Translation for Virtual Antibody-based Tissue Staining Using Local Implicit Functions
May 14, 2025Abstract:Hematoxylin and eosin (H&E) staining is a gold standard for microscopic diagnosis in pathology. However, H&E staining does not capture all the diagnostic information that may be needed. To obtain additional molecular information, immunohistochemical (IHC) stains highlight proteins that mark specific cell types, such as CD3 for T-cells or CK8/18 for epithelial cells. While IHC stains are vital for prognosis and treatment guidance, they are typically only available at specialized centers and time consuming to acquire, leading to treatment delays for patients. Virtual staining, enabled by deep learning-based image translation models, provides a promising alternative by computationally generating IHC stains from H&E stained images. Although many GAN and diffusion based image to image (I2I) translation methods have been used for virtual staining, these models treat image patches as independent data points, which results in increased and more diverse data requirements for effective generation. We present ImplicitStainer, a novel approach that leverages local implicit functions to improve image translation, specifically virtual staining performance, by focusing on pixel-level predictions. This method enhances robustness to variations in dataset sizes, delivering high-quality results even with limited data. We validate our approach on two datasets using a comprehensive set of metrics and benchmark it against over fifteen state-of-the-art GAN- and diffusion based models. Full Code and models trained will be released publicly via Github upon acceptance.
BoundarySeg:An Embarrassingly Simple Method To Boost Medical Image Segmentation Performance for Low Data Regimes
May 14, 2025


Abstract:Obtaining large-scale medical data, annotated or unannotated, is challenging due to stringent privacy regulations and data protection policies. In addition, annotating medical images requires that domain experts manually delineate anatomical structures, making the process both time-consuming and costly. As a result, semi-supervised methods have gained popularity for reducing annotation costs. However, the performance of semi-supervised methods is heavily dependent on the availability of unannotated data, and their effectiveness declines when such data are scarce or absent. To overcome this limitation, we propose a simple, yet effective and computationally efficient approach for medical image segmentation that leverages only existing annotations. We propose BoundarySeg , a multi-task framework that incorporates organ boundary prediction as an auxiliary task to full organ segmentation, leveraging consistency between the two task predictions to provide additional supervision. This strategy improves segmentation accuracy, especially in low data regimes, allowing our method to achieve performance comparable to or exceeding state-of-the-art semi supervised approaches all without relying on unannotated data or increasing computational demands. Code will be released upon acceptance.
Optimization-Driven Statistical Models of Anatomies using Radial Basis Function Shape Representation
Nov 24, 2024



Abstract:Particle-based shape modeling (PSM) is a popular approach to automatically quantify shape variability in populations of anatomies. The PSM family of methods employs optimization to automatically populate a dense set of corresponding particles (as pseudo landmarks) on 3D surfaces to allow subsequent shape analysis. A recent deep learning approach leverages implicit radial basis function representations of shapes to better adapt to the underlying complex geometry of anatomies. Here, we propose an adaptation of this method using a traditional optimization approach that allows more precise control over the desired characteristics of models by leveraging both an eigenshape and a correspondence loss. Furthermore, the proposed approach avoids using a black-box model and allows more freedom for particles to navigate the underlying surfaces, yielding more informative statistical models. We demonstrate the efficacy of the proposed approach to state-of-the-art methods on two real datasets and justify our choice of losses empirically.
VIMs: Virtual Immunohistochemistry Multiplex staining via Text-to-Stain Diffusion Trained on Uniplex Stains
Jul 26, 2024
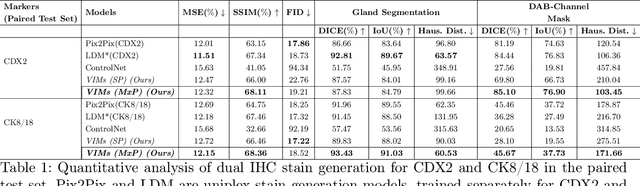

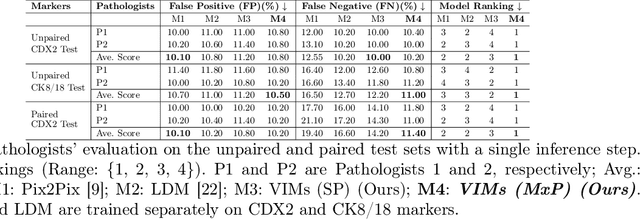
Abstract:This paper introduces a Virtual Immunohistochemistry Multiplex staining (VIMs) model designed to generate multiple immunohistochemistry (IHC) stains from a single hematoxylin and eosin (H&E) stained tissue section. IHC stains are crucial in pathology practice for resolving complex diagnostic questions and guiding patient treatment decisions. While commercial laboratories offer a wide array of up to 400 different antibody-based IHC stains, small biopsies often lack sufficient tissue for multiple stains while preserving material for subsequent molecular testing. This highlights the need for virtual IHC staining. Notably, VIMs is the first model to address this need, leveraging a large vision-language single-step diffusion model for virtual IHC multiplexing through text prompts for each IHC marker. VIMs is trained on uniplex paired H&E and IHC images, employing an adversarial training module. Testing of VIMs includes both paired and unpaired image sets. To enhance computational efficiency, VIMs utilizes a pre-trained large latent diffusion model fine-tuned with small, trainable weights through the Low-Rank Adapter (LoRA) approach. Experiments on nuclear and cytoplasmic IHC markers demonstrate that VIMs outperforms the base diffusion model and achieves performance comparable to Pix2Pix, a standard generative model for paired image translation. Multiple evaluation methods, including assessments by two pathologists, are used to determine the performance of VIMs. Additionally, experiments with different prompts highlight the impact of text conditioning. This paper represents the first attempt to accelerate histopathology research by demonstrating the generation of multiple IHC stains from a single H&E input using a single model trained solely on uniplex data.
Weakly SSM : On the Viability of Weakly Supervised Segmentations for Statistical Shape Modeling
Jul 21, 2024Abstract:Statistical Shape Models (SSMs) excel at identifying population level anatomical variations, which is at the core of various clinical and biomedical applications, including morphology-based diagnostics and surgical planning. However, the effectiveness of SSM is often constrained by the necessity for expert-driven manual segmentation, a process that is both time-intensive and expensive, thereby restricting their broader application and utility. Recent deep learning approaches enable the direct estimation of Statistical Shape Models (SSMs) from unsegmented images. While these models can predict SSMs without segmentation during deployment, they do not address the challenge of acquiring the manual annotations needed for training, particularly in resource-limited settings. Semi-supervised and foundation models for anatomy segmentation can mitigate the annotation burden. Yet, despite the abundance of available approaches, there are no established guidelines to inform end-users on their effectiveness for the downstream task of constructing SSMs. In this study, we systematically evaluate the potential of weakly supervised methods as viable alternatives to manual segmentation's for building SSMs. We establish a new performance benchmark by employing various semi-supervised and foundational model methods for anatomy segmentation under low annotation settings, utilizing the predicted segmentation's for the task of SSM. We compare the modes of shape variation and use quantitative metrics to compare against a shape model derived from a manually annotated dataset. Our results indicate that some methods produce noisy segmentation, which is very unfavorable for SSM tasks, while others can capture the correct modes of variations in the population cohort with 60-80\% reduction in required manual annotation.
Probabilistic 3D Correspondence Prediction from Sparse Unsegmented Images
Jul 02, 2024Abstract:The study of physiology demonstrates that the form (shape)of anatomical structures dictates their functions, and analyzing the form of anatomies plays a crucial role in clinical research. Statistical shape modeling (SSM) is a widely used tool for quantitative analysis of forms of anatomies, aiding in characterizing and identifying differences within a population of subjects. Despite its utility, the conventional SSM construction pipeline is often complex and time-consuming. Additionally, reliance on linearity assumptions further limits the model from capturing clinically relevant variations. Recent advancements in deep learning solutions enable the direct inference of SSM from unsegmented medical images, streamlining the process and improving accessibility. However, the new methods of SSM from images do not adequately account for situations where the imaging data quality is poor or where only sparse information is available. Moreover, quantifying aleatoric uncertainty, which represents inherent data variability, is crucial in deploying deep learning for clinical tasks to ensure reliable model predictions and robust decision-making, especially in challenging imaging conditions. Therefore, we propose SPI-CorrNet, a unified model that predicts 3D correspondences from sparse imaging data. It leverages a teacher network to regularize feature learning and quantifies data-dependent aleatoric uncertainty by adapting the network to predict intrinsic input variances. Experiments on the LGE MRI left atrium dataset and Abdomen CT-1K liver datasets demonstrate that our technique enhances the accuracy and robustness of sparse image-driven SSM.
SCorP: Statistics-Informed Dense Correspondence Prediction Directly from Unsegmented Medical Images
Apr 27, 2024Abstract:Statistical shape modeling (SSM) is a powerful computational framework for quantifying and analyzing the geometric variability of anatomical structures, facilitating advancements in medical research, diagnostics, and treatment planning. Traditional methods for shape modeling from imaging data demand significant manual and computational resources. Additionally, these methods necessitate repeating the entire modeling pipeline to derive shape descriptors (e.g., surface-based point correspondences) for new data. While deep learning approaches have shown promise in streamlining the construction of SSMs on new data, they still rely on traditional techniques to supervise the training of the deep networks. Moreover, the predominant linearity assumption of traditional approaches restricts their efficacy, a limitation also inherited by deep learning models trained using optimized/established correspondences. Consequently, representing complex anatomies becomes challenging. To address these limitations, we introduce SCorP, a novel framework capable of predicting surface-based correspondences directly from unsegmented images. By leveraging the shape prior learned directly from surface meshes in an unsupervised manner, the proposed model eliminates the need for an optimized shape model for training supervision. The strong shape prior acts as a teacher and regularizes the feature learning of the student network to guide it in learning image-based features that are predictive of surface correspondences. The proposed model streamlines the training and inference phases by removing the supervision for the correspondence prediction task while alleviating the linearity assumption.
Estimation and Analysis of Slice Propagation Uncertainty in 3D Anatomy Segmentation
Mar 18, 2024



Abstract:Supervised methods for 3D anatomy segmentation demonstrate superior performance but are often limited by the availability of annotated data. This limitation has led to a growing interest in self-supervised approaches in tandem with the abundance of available un-annotated data. Slice propagation has emerged as an self-supervised approach that leverages slice registration as a self-supervised task to achieve full anatomy segmentation with minimal supervision. This approach significantly reduces the need for domain expertise, time, and the cost associated with building fully annotated datasets required for training segmentation networks. However, this shift toward reduced supervision via deterministic networks raises concerns about the trustworthiness and reliability of predictions, especially when compared with more accurate supervised approaches. To address this concern, we propose the integration of calibrated uncertainty quantification (UQ) into slice propagation methods, providing insights into the model's predictive reliability and confidence levels. Incorporating uncertainty measures enhances user confidence in self-supervised approaches, thereby improving their practical applicability. We conducted experiments on three datasets for 3D abdominal segmentation using five UQ methods. The results illustrate that incorporating UQ improves not only model trustworthiness, but also segmentation accuracy. Furthermore, our analysis reveals various failure modes of slice propagation methods that might not be immediately apparent to end-users. This study opens up new research avenues to improve the accuracy and trustworthiness of slice propagation methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge