Mokshagna Sai Teja Karanam
Mesh2SSM++: A Probabilistic Framework for Unsupervised Learning of Statistical Shape Model of Anatomies from Surface Meshes
Feb 11, 2025
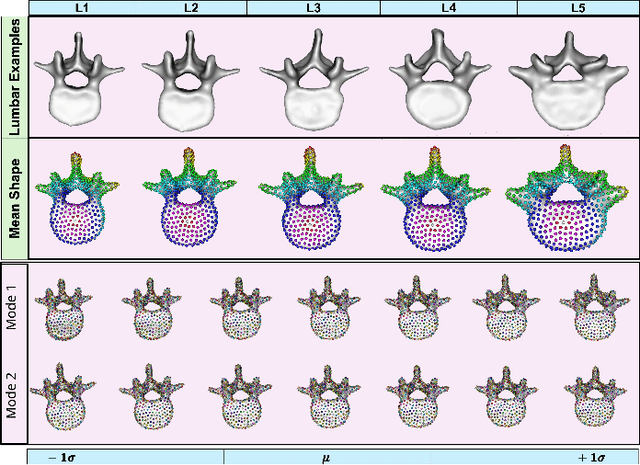
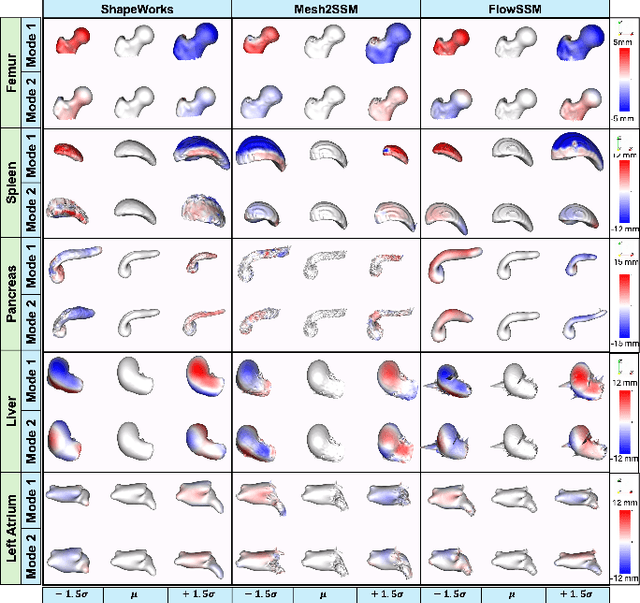
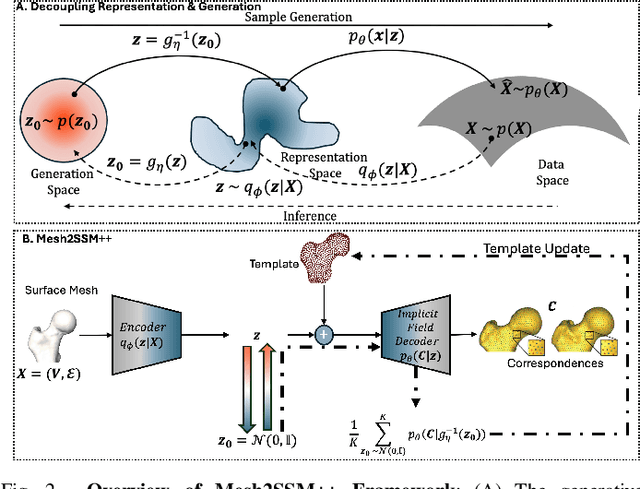
Abstract:Anatomy evaluation is crucial for understanding the physiological state, diagnosing abnormalities, and guiding medical interventions. Statistical shape modeling (SSM) is vital in this process. By enabling the extraction of quantitative morphological shape descriptors from MRI and CT scans, SSM provides comprehensive descriptions of anatomical variations within a population. However, the effectiveness of SSM in anatomy evaluation hinges on the quality and robustness of the shape models. While deep learning techniques show promise in addressing these challenges by learning complex nonlinear representations of shapes, existing models still have limitations and often require pre-established shape models for training. To overcome these issues, we propose Mesh2SSM++, a novel approach that learns to estimate correspondences from meshes in an unsupervised manner. This method leverages unsupervised, permutation-invariant representation learning to estimate how to deform a template point cloud into subject-specific meshes, forming a correspondence-based shape model. Additionally, our probabilistic formulation allows learning a population-specific template, reducing potential biases associated with template selection. A key feature of Mesh2SSM++ is its ability to quantify aleatoric uncertainty, which captures inherent data variability and is essential for ensuring reliable model predictions and robust decision-making in clinical tasks, especially under challenging imaging conditions. Through extensive validation across diverse anatomies, evaluation metrics, and downstream tasks, we demonstrate that Mesh2SSM++ outperforms existing methods. Its ability to operate directly on meshes, combined with computational efficiency and interpretability through its probabilistic framework, makes it an attractive alternative to traditional and deep learning-based SSM approaches.
MORPH-LER: Log-Euclidean Regularization for Population-Aware Image Registration
Feb 04, 2025



Abstract:Spatial transformations that capture population-level morphological statistics are critical for medical image analysis. Commonly used smoothness regularizers for image registration fail to integrate population statistics, leading to anatomically inconsistent transformations. Inverse consistency regularizers promote geometric consistency but lack population morphometrics integration. Regularizers that constrain deformation to low-dimensional manifold methods address this. However, they prioritize reconstruction over interpretability and neglect diffeomorphic properties, such as group composition and inverse consistency. We introduce MORPH-LER, a Log-Euclidean regularization framework for population-aware unsupervised image registration. MORPH-LER learns population morphometrics from spatial transformations to guide and regularize registration networks, ensuring anatomically plausible deformations. It features a bottleneck autoencoder that computes the principal logarithm of deformation fields via iterative square-root predictions. It creates a linearized latent space that respects diffeomorphic properties and enforces inverse consistency. By integrating a registration network with a diffeomorphic autoencoder, MORPH-LER produces smooth, meaningful deformation fields. The framework offers two main contributions: (1) a data-driven regularization strategy that incorporates population-level anatomical statistics to enhance transformation validity and (2) a linearized latent space that enables compact and interpretable deformation fields for efficient population morphometrics analysis. We validate MORPH-LER across two families of deep learning-based registration networks, demonstrating its ability to produce anatomically accurate, computationally efficient, and statistically meaningful transformations on the OASIS-1 brain imaging dataset.
EfficientMorph: Parameter-Efficient Transformer-Based Architecture for 3D Image Registration
Mar 16, 2024



Abstract:Transformers have emerged as the state-of-the-art architecture in medical image registration, outperforming convolutional neural networks (CNNs) by addressing their limited receptive fields and overcoming gradient instability in deeper models. Despite their success, transformer-based models require substantial resources for training, including data, memory, and computational power, which may restrict their applicability for end users with limited resources. In particular, existing transformer-based 3D image registration architectures face three critical gaps that challenge their efficiency and effectiveness. Firstly, while mitigating the quadratic complexity of full attention by focusing on local regions, window-based attention mechanisms often fail to adequately integrate local and global information. Secondly, feature similarities across attention heads that were recently found in multi-head attention architectures indicate a significant computational redundancy, suggesting that the capacity of the network could be better utilized to enhance performance. Lastly, the granularity of tokenization, a key factor in registration accuracy, presents a trade-off; smaller tokens improve detail capture at the cost of higher computational complexity, increased memory demands, and a risk of overfitting. Here, we propose EfficientMorph, a transformer-based architecture for unsupervised 3D image registration. It optimizes the balance between local and global attention through a plane-based attention mechanism, reduces computational redundancy via cascaded group attention, and captures fine details without compromising computational efficiency, thanks to a Hi-Res tokenization strategy complemented by merging operations. Notably, EfficientMorph sets a new benchmark for performance on the OASIS dataset with 16-27x fewer parameters.
ADASSM: Adversarial Data Augmentation in Statistical Shape Models From Images
Jul 10, 2023



Abstract:Statistical shape models (SSM) have been well-established as an excellent tool for identifying variations in the morphology of anatomy across the underlying population. Shape models use consistent shape representation across all the samples in a given cohort, which helps to compare shapes and identify the variations that can detect pathologies and help in formulating treatment plans. In medical imaging, computing these shape representations from CT/MRI scans requires time-intensive preprocessing operations, including but not limited to anatomy segmentation annotations, registration, and texture denoising. Deep learning models have demonstrated exceptional capabilities in learning shape representations directly from volumetric images, giving rise to highly effective and efficient Image-to-SSM. Nevertheless, these models are data-hungry and due to the limited availability of medical data, deep learning models tend to overfit. Offline data augmentation techniques, that use kernel density estimation based (KDE) methods for generating shape-augmented samples, have successfully aided Image-to-SSM networks in achieving comparable accuracy to traditional SSM methods. However, these augmentation methods focus on shape augmentation, whereas deep learning models exhibit image-based texture bias results in sub-optimal models. This paper introduces a novel strategy for on-the-fly data augmentation for the Image-to-SSM framework by leveraging data-dependent noise generation or texture augmentation. The proposed framework is trained as an adversary to the Image-to-SSM network, augmenting diverse and challenging noisy samples. Our approach achieves improved accuracy by encouraging the model to focus on the underlying geometry rather than relying solely on pixel values.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge