Bodong Zhang
DuoFormer: Leveraging Hierarchical Representations by Local and Global Attention Vision Transformer
Jun 15, 2025Abstract:Despite the widespread adoption of transformers in medical applications, the exploration of multi-scale learning through transformers remains limited, while hierarchical representations are considered advantageous for computer-aided medical diagnosis. We propose a novel hierarchical transformer model that adeptly integrates the feature extraction capabilities of Convolutional Neural Networks (CNNs) with the advanced representational potential of Vision Transformers (ViTs). Addressing the lack of inductive biases and dependence on extensive training datasets in ViTs, our model employs a CNN backbone to generate hierarchical visual representations. These representations are adapted for transformer input through an innovative patch tokenization process, preserving the inherited multi-scale inductive biases. We also introduce a scale-wise attention mechanism that directly captures intra-scale and inter-scale associations. This mechanism complements patch-wise attention by enhancing spatial understanding and preserving global perception, which we refer to as local and global attention, respectively. Our model significantly outperforms baseline models in terms of classification accuracy, demonstrating its efficiency in bridging the gap between Convolutional Neural Networks (CNNs) and Vision Transformers (ViTs). The components are designed as plug-and-play for different CNN architectures and can be adapted for multiple applications. The code is available at https://github.com/xiaoyatang/DuoFormer.git.
WeakSupCon: Weakly Supervised Contrastive Learning for Encoder Pre-training
Mar 06, 2025



Abstract:Weakly supervised multiple instance learning (MIL) is a challenging task given that only bag-level labels are provided, while each bag typically contains multiple instances. This topic has been extensively studied in histopathological image analysis, where labels are usually available only at the whole slide image (WSI) level, while each whole slide image can be divided into thousands of small image patches for training. The dominant MIL approaches take fixed patch features as inputs to address computational constraints and ensure model stability. These features are commonly generated by encoders pre-trained on ImageNet, foundation encoders pre-trained on large datasets, or through self-supervised learning on local datasets. While the self-supervised encoder pre-training on the same dataset as downstream MIL tasks helps mitigate domain shift and generate better features, the bag-level labels are not utilized during the process, and the features of patches from different categories may cluster together, reducing classification performance on MIL tasks. Recently, pre-training with supervised contrastive learning (SupCon) has demonstrated superior performance compared to self-supervised contrastive learning and even end-to-end training on traditional image classification tasks. In this paper, we propose a novel encoder pre-training method for downstream MIL tasks called Weakly Supervised Contrastive Learning (WeakSupCon) that utilizes bag-level labels. In our method, we employ multi-task learning and define distinct contrastive learning losses for samples with different bag labels. Our experiments demonstrate that the features generated using WeakSupCon significantly enhance MIL classification performance compared to self-supervised approaches across three datasets.
SRA: A Novel Method to Improve Feature Embedding in Self-supervised Learning for Histopathological Images
Oct 31, 2024



Abstract:Self-supervised learning has become a cornerstone in various areas, particularly histopathological image analysis. Image augmentation plays a crucial role in self-supervised learning, as it generates variations in image samples. However, traditional image augmentation techniques often overlook the unique characteristics of histopathological images. In this paper, we propose a new histopathology-specific image augmentation method called stain reconstruction augmentation (SRA). We integrate our SRA with MoCo v3, a leading model in self-supervised contrastive learning, along with our additional contrastive loss terms, and call the new model SRA-MoCo v3. We demonstrate that our SRA-MoCo v3 always outperforms the standard MoCo v3 across various downstream tasks and achieves comparable or superior performance to other foundation models pre-trained on significantly larger histopathology datasets.
PathMoCo: A Novel Framework to Improve Feature Embedding in Self-supervised Contrastive Learning for Histopathological Images
Oct 23, 2024



Abstract:Self-supervised contrastive learning has become a cornerstone in various areas, particularly histopathological image analysis. Image augmentation plays a crucial role in self-supervised contrastive learning, as it generates variations in image samples. However, traditional image augmentation techniques often overlook the unique characteristics of histopathological images. In this paper, we propose a new histopathology-specific image augmentation method called stain reconstruction augmentation (SRA). We integrate our SRA with MoCo v3, a leading model in self-supervised contrastive learning, along with our additional contrastive loss terms, and call the new model PathMoCo. We demonstrate that our PathMoCo always outperforms the standard MoCo v3 across various downstream tasks and achieves comparable or superior performance to other foundation models pre-trained on significantly larger histopathology datasets.
DuoFormer: Leveraging Hierarchical Visual Representations by Local and Global Attention
Jul 18, 2024



Abstract:We here propose a novel hierarchical transformer model that adeptly integrates the feature extraction capabilities of Convolutional Neural Networks (CNNs) with the advanced representational potential of Vision Transformers (ViTs). Addressing the lack of inductive biases and dependence on extensive training datasets in ViTs, our model employs a CNN backbone to generate hierarchical visual representations. These representations are then adapted for transformer input through an innovative patch tokenization. We also introduce a 'scale attention' mechanism that captures cross-scale dependencies, complementing patch attention to enhance spatial understanding and preserve global perception. Our approach significantly outperforms baseline models on small and medium-sized medical datasets, demonstrating its efficiency and generalizability. The components are designed as plug-and-play for different CNN architectures and can be adapted for multiple applications. The code is available at https://github.com/xiaoyatang/DuoFormer.git.
CLASS-M: Adaptive stain separation-based contrastive learning with pseudo-labeling for histopathological image classification
Jan 04, 2024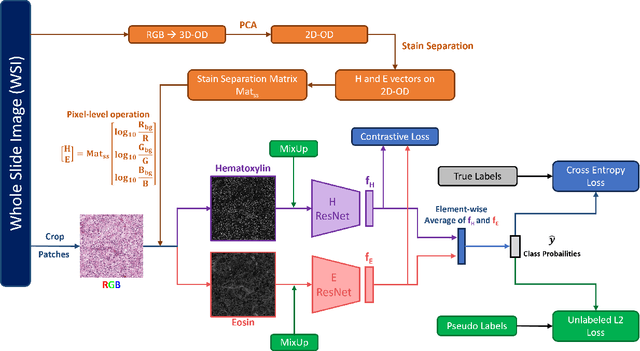
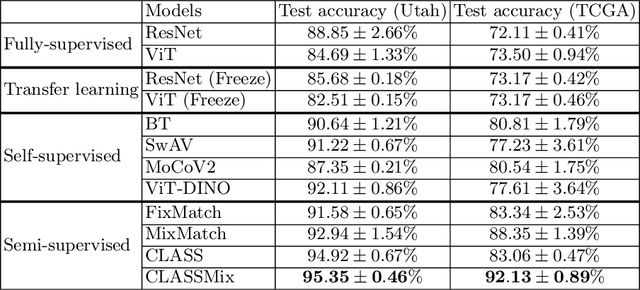
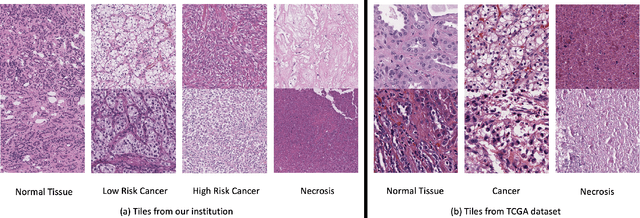
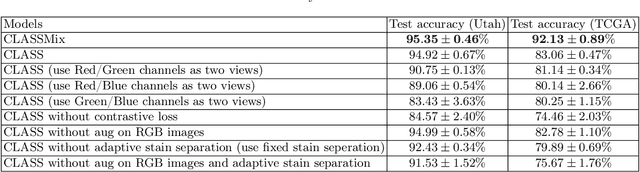
Abstract:Histopathological image classification is an important task in medical image analysis. Recent approaches generally rely on weakly supervised learning due to the ease of acquiring case-level labels from pathology reports. However, patch-level classification is preferable in applications where only a limited number of cases are available or when local prediction accuracy is critical. On the other hand, acquiring extensive datasets with localized labels for training is not feasible. In this paper, we propose a semi-supervised patch-level histopathological image classification model, named CLASS-M, that does not require extensively labeled datasets. CLASS-M is formed by two main parts: a contrastive learning module that uses separated Hematoxylin and Eosin images generated through an adaptive stain separation process, and a module with pseudo-labels using MixUp. We compare our model with other state-of-the-art models on two clear cell renal cell carcinoma datasets. We demonstrate that our CLASS-M model has the best performance on both datasets. Our code is available at github.com/BzhangURU/Paper_CLASS-M/tree/main
Stain based contrastive co-training for histopathological image analysis
Jun 24, 2022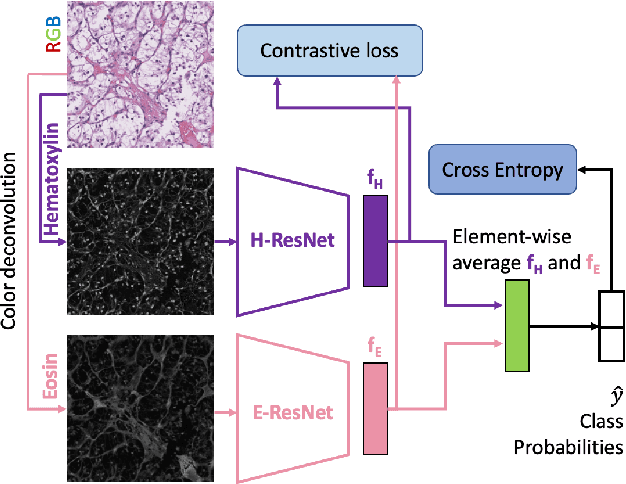
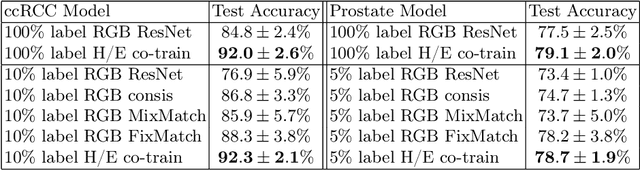
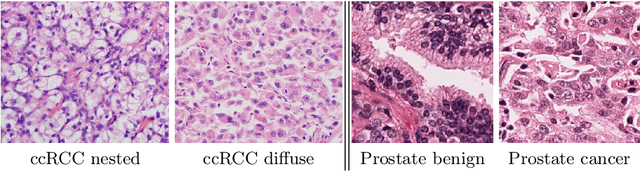

Abstract:We propose a novel semi-supervised learning approach for classification of histopathology images. We employ strong supervision with patch-level annotations combined with a novel co-training loss to create a semi-supervised learning framework. Co-training relies on multiple conditionally independent and sufficient views of the data. We separate the hematoxylin and eosin channels in pathology images using color deconvolution to create two views of each slide that can partially fulfill these requirements. Two separate CNNs are used to embed the two views into a joint feature space. We use a contrastive loss between the views in this feature space to implement co-training. We evaluate our approach in clear cell renal cell and prostate carcinomas, and demonstrate improvement over state-of-the-art semi-supervised learning methods.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge