Wenzheng Tao
Learning Population-level Shape Statistics and Anatomy Segmentation From Images: A Joint Deep Learning Model
Jan 10, 2022



Abstract:Statistical shape modeling is an essential tool for the quantitative analysis of anatomical populations. Point distribution models (PDMs) represent the anatomical surface via a dense set of correspondences, an intuitive and easy-to-use shape representation for subsequent applications. These correspondences are exhibited in two coordinate spaces: the local coordinates describing the geometrical features of each individual anatomical surface and the world coordinates representing the population-level statistical shape information after removing global alignment differences across samples in the given cohort. We propose a deep-learning-based framework that simultaneously learns these two coordinate spaces directly from the volumetric images. The proposed joint model serves a dual purpose; the world correspondences can directly be used for shape analysis applications, circumventing the heavy pre-processing and segmentation involved in traditional PDM models. Additionally, the local correspondences can be used for anatomy segmentation. We demonstrate the efficacy of this joint model for both shape modeling applications on two datasets and its utility in inferring the anatomical surface.
DeepSSM: A Blueprint for Image-to-Shape Deep Learning Models
Oct 14, 2021

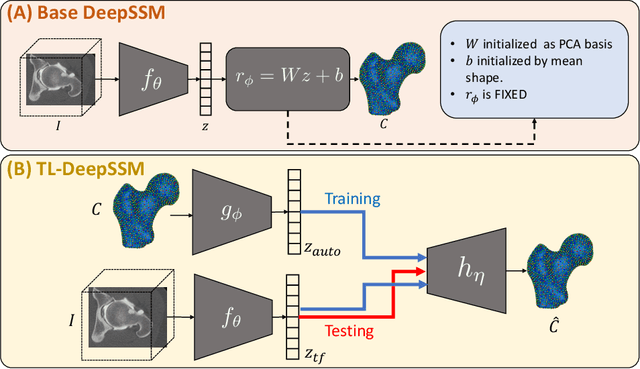
Abstract:Statistical shape modeling (SSM) characterizes anatomical variations in a population of shapes generated from medical images. SSM requires consistent shape representation across samples in shape cohort. Establishing this representation entails a processing pipeline that includes anatomy segmentation, re-sampling, registration, and non-linear optimization. These shape representations are then used to extract low-dimensional shape descriptors that facilitate subsequent analyses in different applications. However, the current process of obtaining these shape descriptors from imaging data relies on human and computational resources, requiring domain expertise for segmenting anatomies of interest. Moreover, this same taxing pipeline needs to be repeated to infer shape descriptors for new image data using a pre-trained/existing shape model. Here, we propose DeepSSM, a deep learning-based framework for learning the functional mapping from images to low-dimensional shape descriptors and their associated shape representations, thereby inferring statistical representation of anatomy directly from 3D images. Once trained using an existing shape model, DeepSSM circumvents the heavy and manual pre-processing and segmentation and significantly improves the computational time, making it a viable solution for fully end-to-end SSM applications. In addition, we introduce a model-based data-augmentation strategy to address data scarcity. Finally, this paper presents and analyzes two different architectural variants of DeepSSM with different loss functions using three medical datasets and their downstream clinical application. Experiments showcase that DeepSSM performs comparably or better to the state-of-the-art SSM both quantitatively and on application-driven downstream tasks. Therefore, DeepSSM aims to provide a comprehensive blueprint for deep learning-based image-to-shape models.
Review Regularized Neural Collaborative Filtering
Aug 20, 2020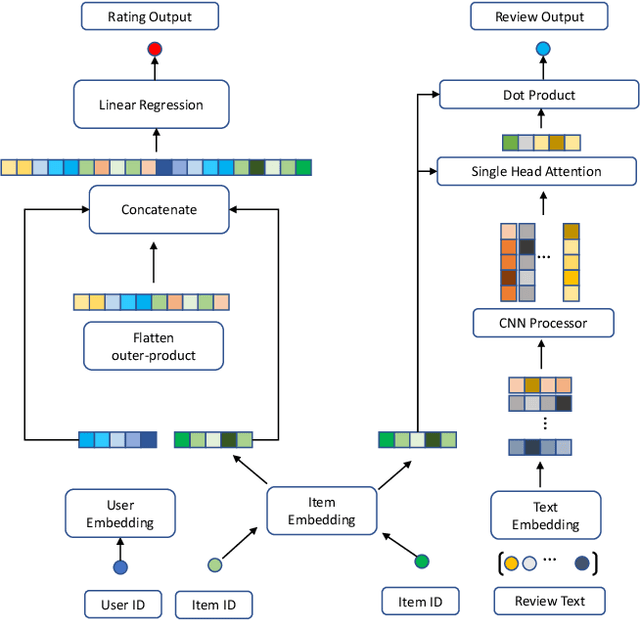
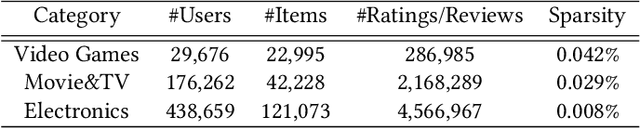
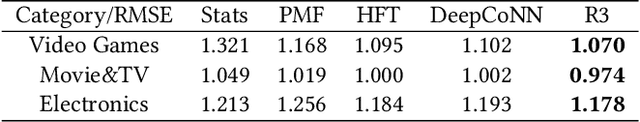

Abstract:In recent years, text-aware collaborative filtering methods have been proposed to address essential challenges in recommendations such as data sparsity, cold start problem, and long-tail distribution. However, many of these text-oriented methods rely heavily on the availability of text information for every user and item, which obviously does not hold in real-world scenarios. Furthermore, specially designed network structures for text processing are highly inefficient for on-line serving and are hard to integrate into current systems. In this paper, we propose a flexible neural recommendation framework, named Review Regularized Recommendation, short as R3. It consists of a neural collaborative filtering part that focuses on prediction output, and a text processing part that serves as a regularizer. This modular design incorporates text information as richer data sources in the training phase while being highly friendly for on-line serving as it needs no on-the-fly text processing in serving time. Our preliminary results show that by using a simple text processing approach, it could achieve better prediction performance than state-of-the-art text-aware methods.
Unsupervised Shape Normality Metric for Severity Quantification
Jul 18, 2020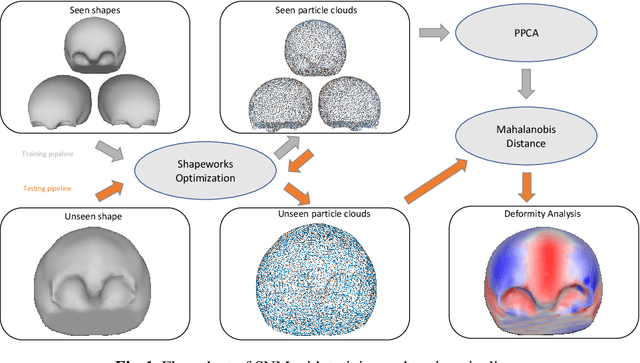
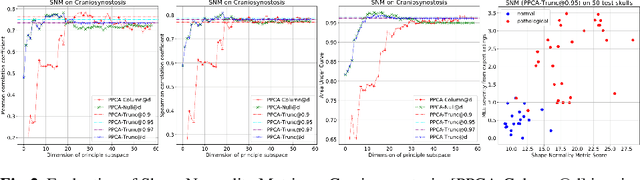

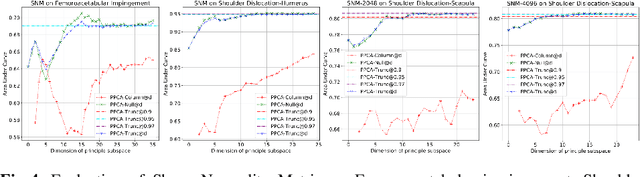
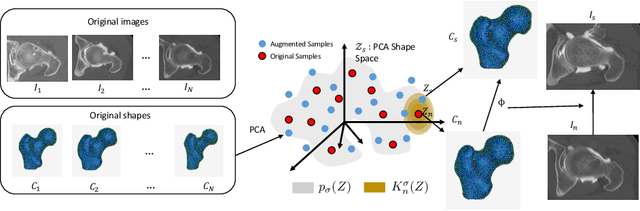
Abstract:This work describes an unsupervised method to objectively quantify the abnormality of general anatomical shapes. The severity of an anatomical deformity often serves as a determinant in the clinical management of patients. However, experiential bias and distinctive random residuals among specialist individuals bring variability in diagnosis and patient management decisions, irrespective of the objective deformity degree. Therefore, supervised methods are prone to be misled given insufficient labeling of pathological samples that inevitably preserve human bias and inconsistency. Furthermore, subjects demonstrating a specific pathology are naturally rare relative to the normal population. To avoid relying on sufficient pathological samples by fully utilizing the power of normal samples, we propose the shape normality metric (SNM), which requires learning only from normal samples and zero knowledge about the pathology. We represent shapes by landmarks automatically inferred from the data and model the normal group by a multivariate Gaussian distribution. Extensive experiments on different anatomical datasets, including skulls, femurs, scapulae, and humeri, demonstrate that SNM can provide an effective normality measurement, which can significantly detect and indicate pathology. Therefore, SNM offers promising value in a variety of clinical applications.
 Add to Chrome
Add to Chrome Add to Firefox
Add to Firefox Add to Edge
Add to Edge